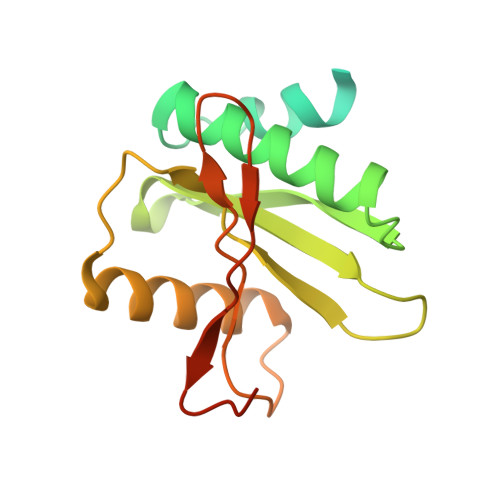
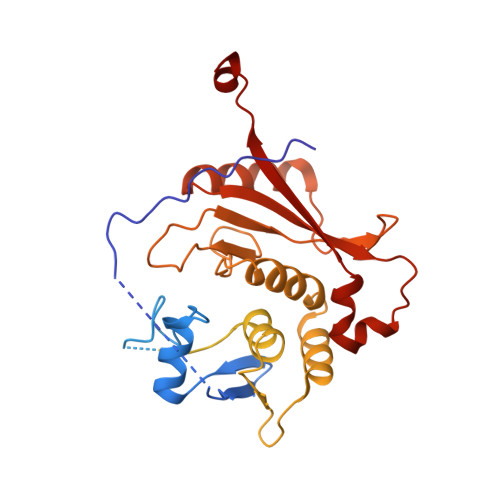
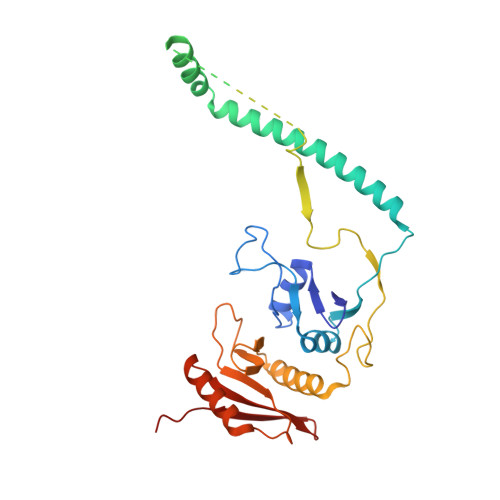
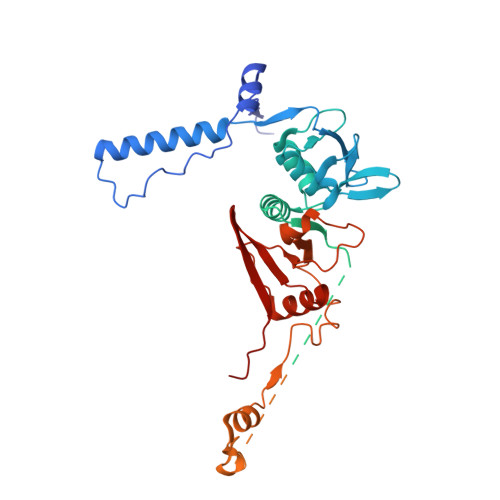
Recognition and cleavage mechanism of intron-containing pre-tRNA by human TSEN endonuclease complex.
Yuan, L., Han, Y., Zhao, J., Zhang, Y., Sun, Y.(2023) Nat Commun 14: 6071-6071
- PubMed: 37770519
- DOI: https://doi.org/10.1038/s41467-023-41845-y
- Primary Citation of Related Structures:
8ISS - PubMed Abstract:
Removal of introns from transfer RNA precursors (pre-tRNAs) occurs in all living organisms. This is a vital phase in the maturation and functionality of tRNA. Here we present a 3.2 Å-resolution cryo-EM structure of an active human tRNA splicing endonuclease complex bound to an intron-containing pre-tRNA. TSEN54, along with the unique regions of TSEN34 and TSEN2, cooperatively recognizes the mature body of pre-tRNA and guides the anticodon-intron stem to the correct position for splicing. We capture the moment when the endonucleases are poised for cleavage, illuminating the molecular mechanism for both 3' and 5' cleavage reactions. Two insertion loops from TSEN54 and TSEN2 cover the 3' and 5' splice sites, respectively, trapping the scissile phosphate in the center of the catalytic triad of residues. Our findings reveal the molecular mechanism for eukaryotic pre-tRNA recognition and cleavage, as well as the evolutionary relationship between archaeal and eukaryotic TSENs.
- School of Life Science and Technology, ShanghaiTech University, Shanghai, China.
Organizational Affiliation: