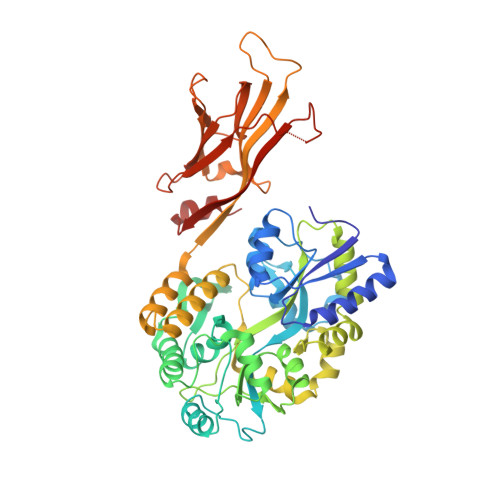
GAS41 promotes H2A.Z deposition through recognition of the N terminus of histone H3 by the YEATS domain.
Kikuchi, M., Takase, S., Konuma, T., Noritsugu, K., Sekine, S., Ikegami, T., Ito, A., Umehara, T.(2023) Proc Natl Acad Sci U S A 120: e2304103120-e2304103120
- PubMed: 37844223
- DOI: https://doi.org/10.1073/pnas.2304103120
- Primary Citation of Related Structures:
7EIF, 8IIY, 8IIZ, 8IJ0 - PubMed Abstract:
Glioma amplified sequence 41 (GAS41), which has the Yaf9, ENL, AF9, Taf14, and Sas5 (YEATS) domain that recognizes lysine acetylation (Kac), regulates gene expression as a subunit of the SRCAP (SNF2-related CREBBP activator protein) complex that deposits histone H2A.Z at promoters in eukaryotes. The YEATS domains of the proteins AF9 and ENL recognize Kac by hydrogen bonding the aromatic cage to arginine situated just before K9ac or K27ac in the N-terminal tail of histone H3. Curiously, the YEATS domain of GAS41 binds most preferentially to the sequence that contains K14ac of H3 (H3K14ac) but lacks the corresponding arginine. Here, we biochemically and structurally elucidated the molecular mechanism by which GAS41 recognizes H3K14ac. First, stable binding of the GAS41 YEATS domain to H3K14ac required the N terminus of H3 (H3NT). Second, we revealed a pocket in the GAS41 YEATS domain responsible for the H3NT binding by crystallographic and NMR analyses. This pocket is away from the aromatic cage that recognizes Kac and is unique to GAS41 among the YEATS family. Finally, we showed that E109 of GAS41, a residue essential for the formation of the H3NT-binding pocket, was crucial for chromatin occupancy of H2A.Z and GAS41 at H2A.Z-enriched promoter regions. These data suggest that binding of GAS41 to H3NT via its YEATS domain is essential for its intracellular function.
- Laboratory for Epigenetics Drug Discovery, RIKEN Center for Biosystems Dynamics Research, Yokohama 230-0045, Japan.
Organizational Affiliation: