Structure-Based Design of Potent Peptidomimetic Inhibitors Covalently Targeting SARS-CoV-2 Papain-like Protease.
Wang, Q., Chen, G., He, J., Li, J., Xiong, M., Su, H., Li, M., Hu, H., Xu, Y.(2023) Int J Mol Sci 24
- PubMed: 37239980
- DOI: https://doi.org/10.3390/ijms24108633
- Primary Citation of Related Structures:
8IHO - PubMed Abstract:
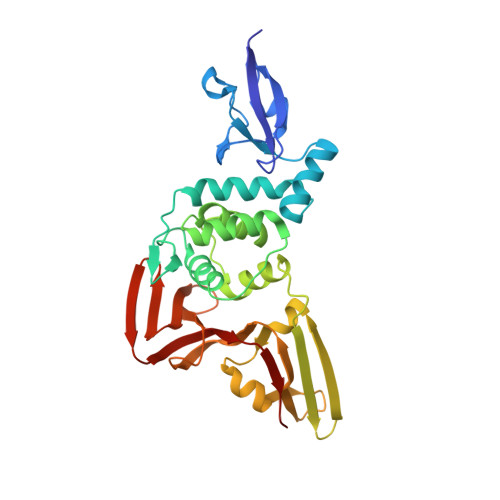
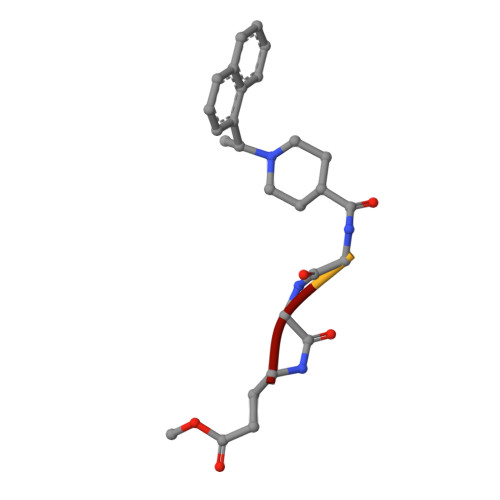
The papain-like protease (PL pro ) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) plays a critical role in the proteolytic processing of viral polyproteins and the dysregulation of the host immune response, providing a promising therapeutic target. Here, we report the structure-guide design of novel peptidomimetic inhibitors covalently targeting SARS-CoV-2 PL pro . The resulting inhibitors demonstrate submicromolar potency in the enzymatic assay (IC 50 = 0.23 μM) and significant inhibition of SARS-CoV-2 PL pro in the HEK293T cells using a cell-based protease assay (EC 50 = 3.61 μM). Moreover, an X-ray crystal structure of SARS-CoV-2 PL pro in complex with compound 2 confirms the covalent binding of the inhibitor to the catalytic residue cysteine 111 (C111) and emphasizes the importance of interactions with tyrosine 268 (Y268). Together, our findings reveal a new scaffold of SARS-CoV-2 PL pro inhibitors and provide an attractive starting point for further optimization.
- School of Chinese Materia Medica, Nanjing University of Chinese Medicine, Nanjing 210023, China.
Organizational Affiliation: