The mechanism of DRB7.2:DRB4 mediated sequestering of endogenous inverted-repeat dsRNA precursors in plants
Paturi, S., Patra, D., Behera, P.C., Aute, R., Waghela, N., Kinatukara, P., Deshmukh, M.V.(2025) Elife
Experimental Data Snapshot
Starting Model: in silico
View more details
wwPDB Validation 3D Report Full Report
(2025) Elife
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Double-stranded RNA-binding domain (DsRBD)-containing protein | 97 | Arabidopsis thaliana | Mutation(s): 0 Gene Names: AT4G00420 | 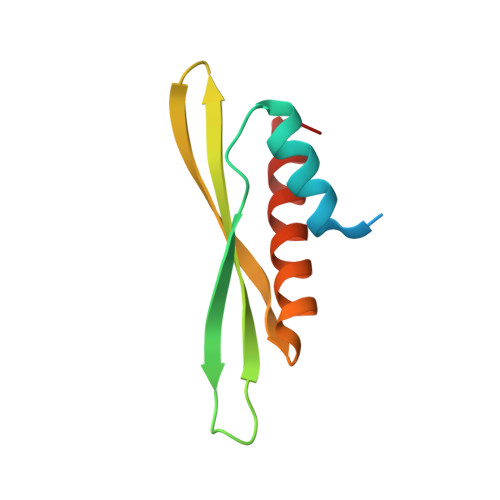 | |
UniProt | |||||
Find proteins for F4JHB3 (Arabidopsis thaliana) Explore F4JHB3 Go to UniProtKB: F4JHB3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | F4JHB3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Double-stranded RNA-binding protein 4 | 71 | Arabidopsis thaliana | Mutation(s): 0 Gene Names: DBR4 | 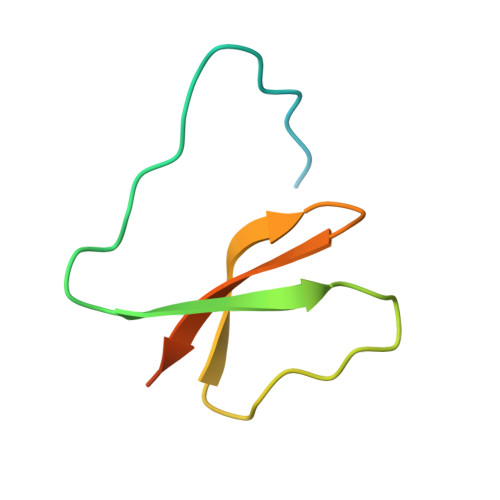 | |
UniProt | |||||
Find proteins for Q8H1D4 (Arabidopsis thaliana) Explore Q8H1D4 Go to UniProtKB: Q8H1D4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8H1D4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 93.94 | α = 90 |
| b = 93.94 | β = 90 |
| c = 102.81 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| iMOSFLM | data reduction |
| SCALA | data scaling |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Council of Scientific & Industrial Research (CSIR) | India | MLP0161 |