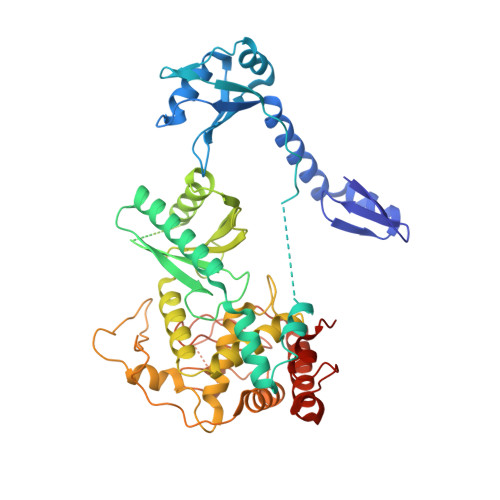
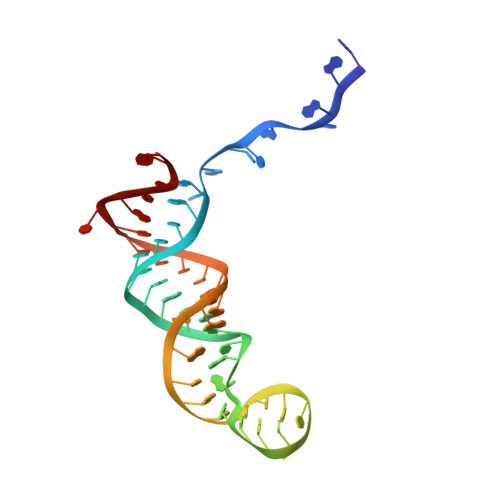
Mechanism of U6 snRNA oligouridylation by human TUT1.
Yamashita, S., Tomita, K.(2023) Nat Commun 14: 4686-4686
- PubMed: 37563152
- DOI: https://doi.org/10.1038/s41467-023-40420-9
- Primary Citation of Related Structures:
8IDF - PubMed Abstract:
U6 snRNA is a catalytic RNA responsible for pre-mRNA splicing reactions and undergoes various post-transcriptional modifications during its maturation process. The 3'-oligouridylation of U6 snRNA by the terminal uridylyltransferase, TUT1, provides the Lsm-binding site in U6 snRNA for U4/U6 di-snRNP formation and this ensures pre-mRNA splicing. Here, we present the crystal structure of human TUT1 (hTUT1) complexed with U6 snRNA, representing the post-uridylation of U6 snRNA by hTUT1. The N-terminal ZF-RRM and catalytic palm clamp the single-stranded AUA motif between the 5'-short stem and the 3'-telestem of U6 snRNA, and the ZF-RRM specifically recognizes the AUA motif. The ZF and the fingers hold the telestem, and the 3'-end of U6 snRNA is placed in the catalytic pocket of the palm for oligouridylation. The oligouridylation of U6 snRNA depends on the internal four-adenosine tract in the 5'-part of the telestem of U6 snRNA, and hTUT1 adds uridines until the internal adenosine tract can form base-pairs with the 3'-oligouridine tract. Together, the recognition of the specific structure and sequence of U6 snRNA by the multi-domain TUT1 protein and the intrinsic sequence and structure of U6 snRNA ensure the oligouridylation of U6 snRNA.
- Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Kashiwa, Chiba, 277-8562, Japan.
Organizational Affiliation: