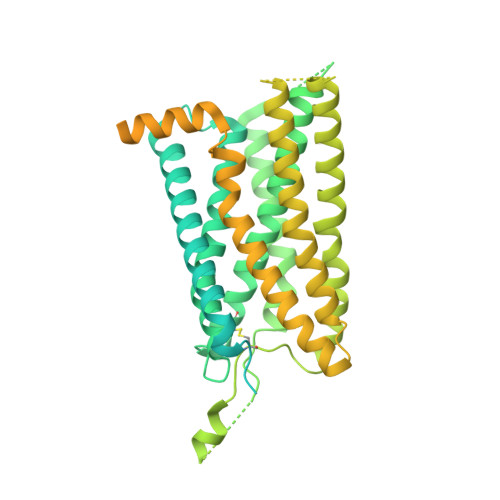
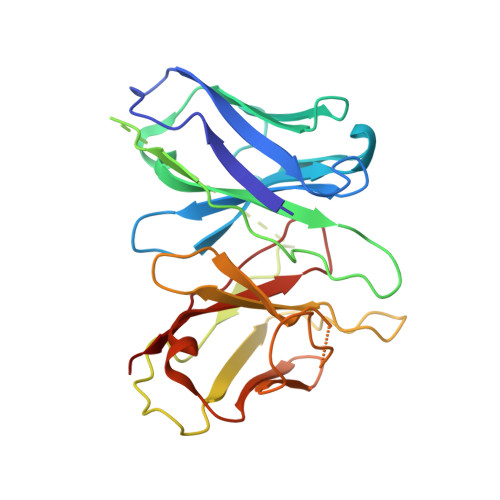
Structural basis for motilin and erythromycin recognition by motilin receptor.
You, C., Zhang, Y., Xu, Y., Xu, P., Li, Z., Li, H., Huang, S., Chen, Z., Li, J., Xu, H.E., Jiang, Y.(2023) Sci Adv 9: eade9020-eade9020
- PubMed: 36921049
- DOI: https://doi.org/10.1126/sciadv.ade9020
- Primary Citation of Related Structures:
8IBU, 8IBV - PubMed Abstract:
Motilin is an endogenous peptide hormone almost exclusively expressed in the human gastrointestinal (GI) tract. It activates the motilin receptor (MTLR), a class A G protein-coupled receptor (GPCR), and stimulates GI motility. To our knowledge, MTLR is the first GPCR reported to be activated by macrolide antibiotics, such as erythromycin. It has attracted extensive attention as a potential drug target for GI disorders. We report two structures of G q -coupled human MTLR bound to motilin and erythromycin. Our structures reveal the recognition mechanism of both ligands and explain the specificity of motilin and ghrelin, a related gut peptide hormone, for their respective receptors. These structures also provide the basis for understanding the different recognition modes of erythromycin by MTLR and ribosome. These findings provide a framework for understanding the physiological regulation of MTLR and guiding drug design targeting MTLR for the treatment of GI motility disorders.
- State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China.
Organizational Affiliation: