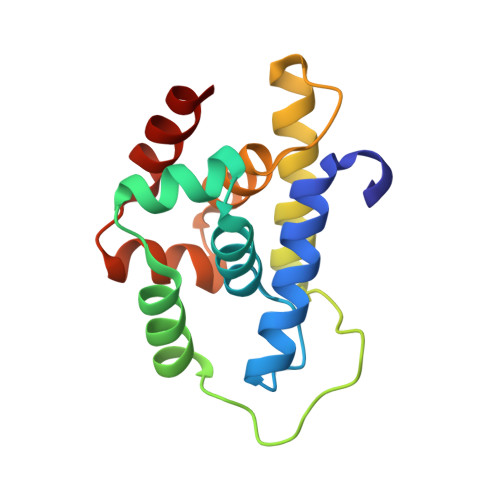
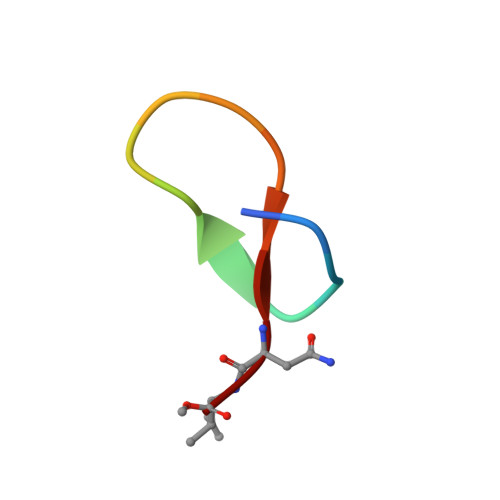
Crystal structure of the N-terminal domain of MtClpC1 in complex with the anti-mycobacterial natural peptide Lassomycin.
Jagdev, M.K., Tompa, D.R., Ling, L.L., Peoples, A.J., Dandapat, J., Mohapatra, C., Lewis, K., Vasudevan, D.(2023) Int J Biol Macromol 253: 126771-126771
- PubMed: 37683752
- DOI: https://doi.org/10.1016/j.ijbiomac.2023.126771
- Primary Citation of Related Structures:
8IBO, 8IBP - PubMed Abstract:
Antibiotics form our frontline therapy against disease-causing bacteria. Unfortunately, antibiotic resistance is becoming more common, threatening a future where these medications can no longer cure infections. Furthermore, the emergence of multidrug-resistant (MDR), totally drug-resistant (TDR), and extensively drug-resistant (XDR) tuberculosis has increased the urgency of discovering new therapeutic leads with unique modes of action. Some natural peptides derived from actinomycetes, such as Cyclomarin A, Lassomycin, Rufomycin I, and Ecumicin, have potent and specific bactericidal activity against Mycobacterium tuberculosis, with the specificity owing to the fact that these peptides target the ClpC1 ATPase, an essential enzyme in mycobacteria, and inhibit/activate the proteolytic activity of the ClpC1/P1/P2 complex that participates in protein homeostasis. Here, we report the high-resolution crystal structure of the N-terminal domain of ClpC1 (ClpC1 NTD) in complex with Lassomycin, showing the specific binding mode of Lassomycin. In addition, the work also compares the Lassomycin complex structure with the previously known structures of ClpC1 NTD in complex with other natural peptides such as Cyclomarin A, Rufomycin I, and Ecumicin.
- Division of Infectious Disease Biology, Institute of Life Sciences, Bhubaneswar 751023, Odisha, India; Post-Graduate Department of Biotechnology, Utkal University, Bhubaneswar 751004, Odisha, India.
Organizational Affiliation: