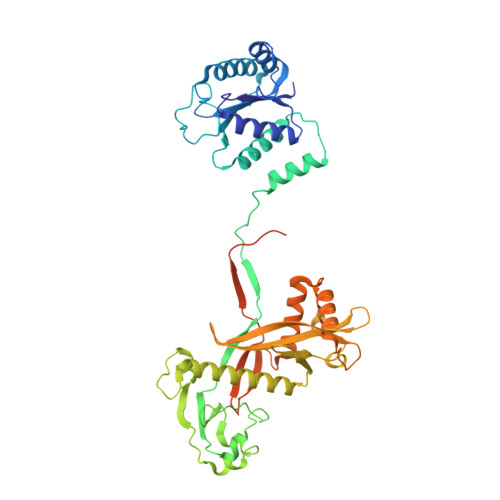
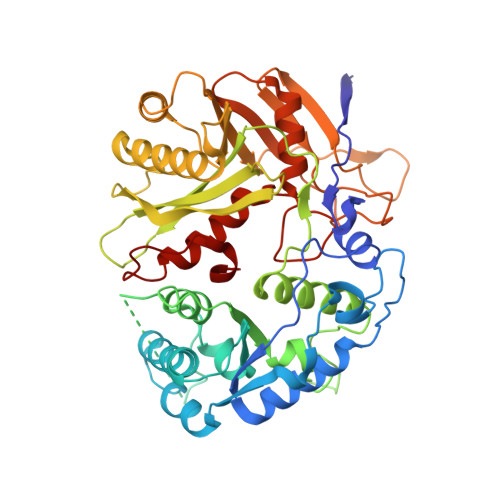
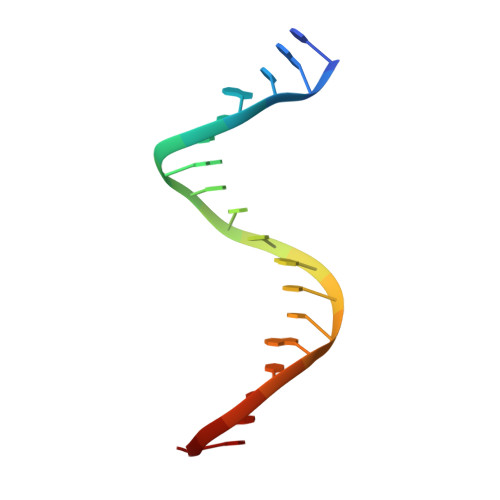
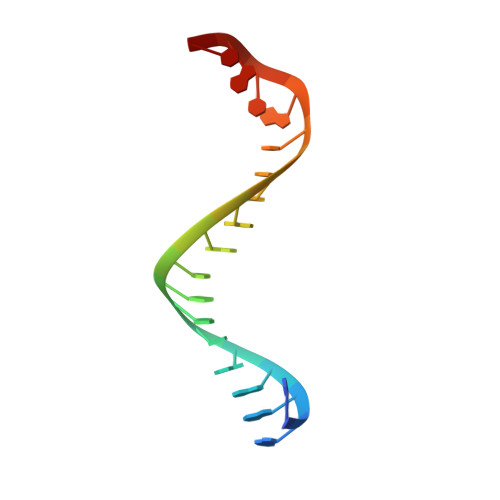
Structural insights into mechanisms of Argonaute protein-associated NADase activation in bacterial immunity.
Wang, X., Li, X., Yu, G., Zhang, L., Zhang, C., Wang, Y., Liao, F., Wen, Y., Yin, H., Liu, X., Wei, Y., Li, Z., Deng, Z., Zhang, H.(2023) Cell Res 33: 699-711
- PubMed: 37311833
- DOI: https://doi.org/10.1038/s41422-023-00839-7
- Primary Citation of Related Structures:
8I87, 8I88, 8IN8 - PubMed Abstract:
Nicotinamide adenine dinucleotide (NAD + ) is a central metabolite in cellular processes. Depletion of NAD + has been demonstrated to be a prevalent theme in both prokaryotic and eukaryotic immune responses. Short prokaryotic Argonaute proteins (Agos) are associated with NADase domain-containing proteins (TIR-APAZ or SIR2-APAZ) encoded in the same operon. They confer immunity against mobile genetic elements, such as bacteriophages and plasmids, by inducing NAD + depletion upon recognition of target nucleic acids. However, the molecular mechanisms underlying the activation of such prokaryotic NADase/Ago immune systems remain unknown. Here, we report multiple cryo-EM structures of NADase/Ago complexes from two distinct systems (TIR-APAZ/Ago and SIR2-APAZ/Ago). Target DNA binding triggers tetramerization of the TIR-APAZ/Ago complex by a cooperative self-assembly mechanism, while the heterodimeric SIR2-APAZ/Ago complex does not assemble into higher-order oligomers upon target DNA binding. However, the NADase activities of these two systems are unleashed via a similar closed-to-open transition of the catalytic pocket, albeit by different mechanisms. Furthermore, a functionally conserved sensor loop is employed to inspect the guide RNA-target DNA base pairing and facilitate the conformational rearrangement of Ago proteins required for the activation of these two systems. Overall, our study reveals the mechanistic diversity and similarity of Ago protein-associated NADase systems in prokaryotic immune response.
- National key laboratory of blood science, The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), Haihe Laboratory of Cell Ecosystem, Tianjin Institute of Immunology, Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University, Tianjin, China.
Organizational Affiliation: