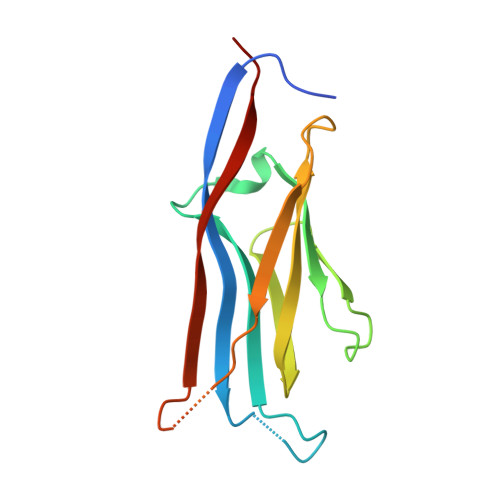
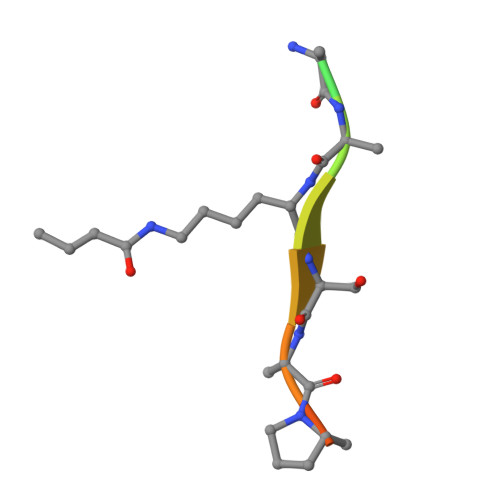
Histone H3 lysine 27 crotonylation mediates gene transcriptional repression in chromatin.
Liu, N., Konuma, T., Sharma, R., Wang, D., Zhao, N., Cao, L., Ju, Y., Liu, D., Wang, S., Bosch, A., Sun, Y., Zhang, S., Ji, D., Nagatoishi, S., Suzuki, N., Kikuchi, M., Wakamori, M., Zhao, C., Ren, C., Zhou, T.J., Xu, Y., Meslamani, J., Fu, S., Umehara, T., Tsumoto, K., Akashi, S., Zeng, L., Roeder, R.G., Walsh, M.J., Zhang, Q., Zhou, M.M.(2023) Mol Cell 83: 2206-2221.e11
- PubMed: 37311463
- DOI: https://doi.org/10.1016/j.molcel.2023.05.022
- Primary Citation of Related Structures:
8I60 - PubMed Abstract:
Histone lysine acylation, including acetylation and crotonylation, plays a pivotal role in gene transcription in health and diseases. However, our understanding of histone lysine acylation has been limited to gene transcriptional activation. Here, we report that histone H3 lysine 27 crotonylation (H3K27cr) directs gene transcriptional repression rather than activation. Specifically, H3K27cr in chromatin is selectively recognized by the YEATS domain of GAS41 in complex with SIN3A-HDAC1 co-repressors. Proto-oncogenic transcription factor MYC recruits GAS41/SIN3A-HDAC1 complex to repress genes in chromatin, including cell-cycle inhibitor p21. GAS41 knockout or H3K27cr-binding depletion results in p21 de-repression, cell-cycle arrest, and tumor growth inhibition in mice, explaining a causal relationship between GAS41 and MYC gene amplification and p21 downregulation in colorectal cancer. Our study suggests that H3K27 crotonylation signifies a previously unrecognized, distinct chromatin state for gene transcriptional repression in contrast to H3K27 trimethylation for transcriptional silencing and H3K27 acetylation for transcriptional activation.
- Bethune Institute of Epigenetic Medicine, First Hospital of Jilin University, Changchun 130061, China; International Center of Future Science, Jilin University, Changchun 130012, China. Electronic address: nanliu@mail.jlu.edu.cn.
Organizational Affiliation: