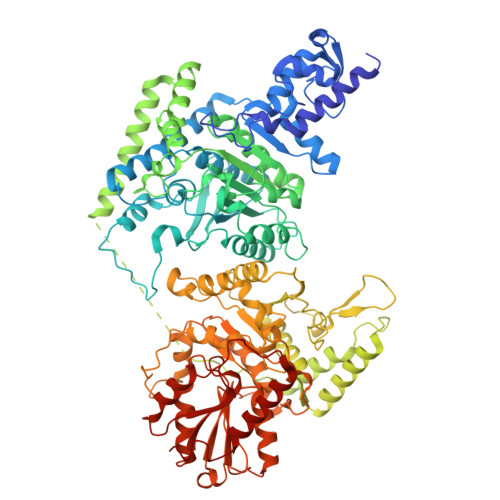
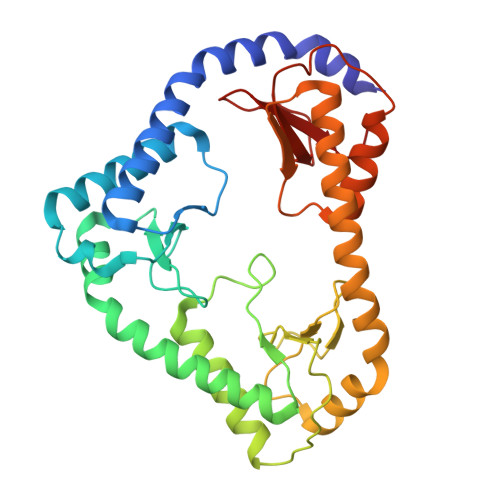
The mitochondrial DNAJC co-chaperone TCAIM reduces alpha-ketoglutarate dehydrogenase protein levels to regulate metabolism
Wang, J., Yu, X., Zhong, Y., Ma, X., Gao, Y., Zhou, D., Wang, J., Fu, Y., Fan, S., Su, J., Huang, M., Haigis, M., Wang, P., Xu, Y., Yang, W.(2025) Mol Cell 85: 638