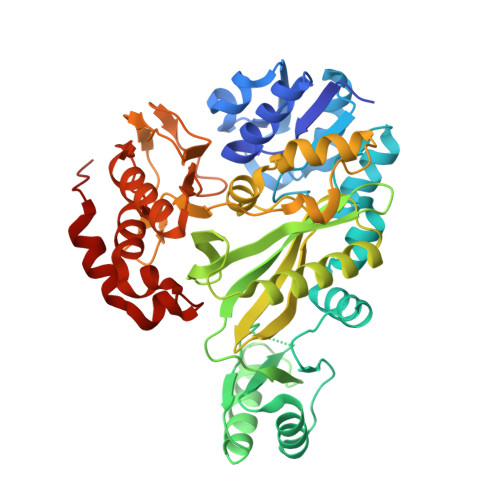
Chloroflexus aurantiacus acetyl-CoA carboxylase evolves fused biotin carboxylase and biotin carboxyl carrier protein to complete carboxylation activity.
Shen, J., Wu, W., Wang, K., Wu, J., Liu, B., Li, C., Gong, Z., Hong, X., Fang, H., Zhang, X., Xu, X.(2024) mBio 15: e0341423-e0341423
- PubMed: 38572988
- DOI: https://doi.org/10.1128/mbio.03414-23
- Primary Citation of Related Structures:
8HZ4, 8HZ5 - PubMed Abstract:
Acetyl-CoA carboxylases (ACCs) convert acetyl-CoA to malonyl-CoA, a key step in fatty acid biosynthesis and autotrophic carbon fixation pathways. Three functionally distinct components, biotin carboxylase (BC), biotin carboxyl carrier protein (BCCP), and carboxyltransferase (CT), are either separated or partially fused in different combinations, forming heteromeric ACCs. However, an ACC with fused BC-BCCP and separate CT has not been identified, leaving its catalytic mechanism unclear. Here, we identify two BC isoforms (BC1 and BC2) from Chloroflexus aurantiacus , a filamentous anoxygenic phototroph that employs 3-hydroxypropionate (3-HP) bi-cycle rather than Calvin cycle for autotrophic carbon fixation. We reveal that BC1 possesses fused BC and BCCP domains, where BCCP could be biotinylated by E. coli or C. aurantiacus BirA on Lys553 residue. Crystal structures of BC1 and BC2 at 3.2 Å and 3.0 Å resolutions, respectively, further reveal a tetramer of two BC1-BC homodimers, and a BC2 homodimer, all exhibiting similar BC architectures. The two BC1-BC homodimers are connected by an eight-stranded β-barrel of the partially resolved BCCP domain. Disruption of β-barrel results in dissociation of the tetramer into dimers in solution and decreased biotin carboxylase activity. Biotinylation of the BCCP domain further promotes BC1 and CTβ-CTα interactions to form an enzymatically active ACC, which converts acetyl-CoA to malonyl-CoA in vitro and produces 3-HP via co-expression with a recombinant malonyl-CoA reductase in E. coli cells. This study revealed a heteromeric ACC that evolves fused BC-BCCP but separate CTα and CTβ to complete ACC activity.IMPORTANCEAcetyl-CoA carboxylase (ACC) catalyzes the rate-limiting step in fatty acid biosynthesis and autotrophic carbon fixation pathways across a wide range of organisms, making them attractive targets for drug discovery against various infections and diseases. Although structural studies on homomeric ACCs, which consist of a single protein with three subunits, have revealed the "swing domain model" where the biotin carboxyl carrier protein (BCCP) domain translocates between biotin carboxylase (BC) and carboxyltransferase (CT) active sites to facilitate the reaction, our understanding of the subunit composition and catalytic mechanism in heteromeric ACCs remains limited. Here, we identify a novel ACC from an ancient anoxygenic photosynthetic bacterium Chloroflexus aurantiacus , it evolves fused BC and BCCP domain, but separate CT components to form an enzymatically active ACC, which converts acetyl-CoA to malonyl-CoA in vitro and produces 3-hydroxypropionate (3-HP) via co-expression with recombinant malonyl-CoA reductase in E. coli cells. These findings expand the diversity and molecular evolution of heteromeric ACCs and provide a structural basis for potential applications in 3-HP biosynthesis.
- Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Hangzhou Normal University, Hangzhou, China.
Organizational Affiliation: