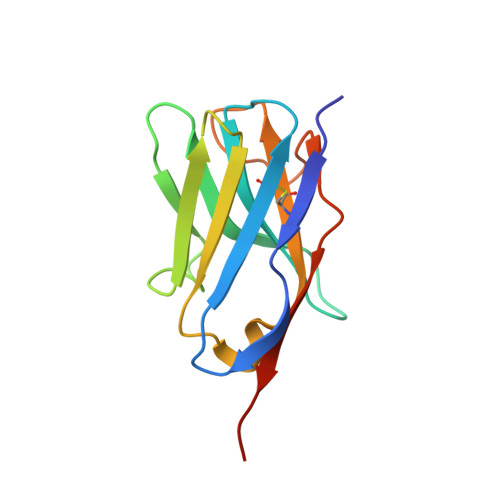
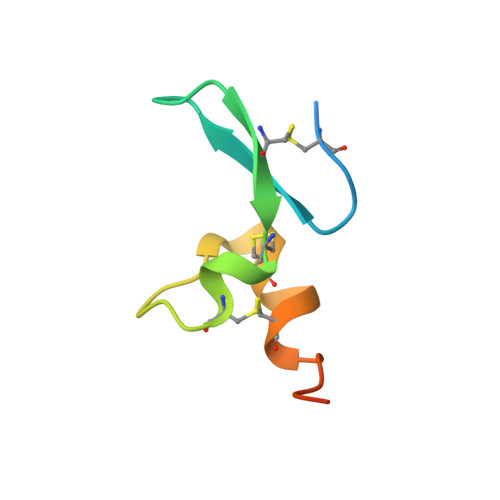
Antigen-induced chimeric antigen receptor multimerization amplifies on-tumor cytotoxicity.
Sun, Y., Yang, X.N., Yang, S.S., Lyu, Y.Z., Zhang, B., Liu, K.W., Li, N., Cui, J.C., Huang, G.X., Liu, C.L., Xu, J., Mi, J.Q., Chen, Z., Fan, X.H., Chen, S.J., Chen, S.(2023) Signal Transduct Target Ther 8: 445-445
- PubMed: 38062078
- DOI: https://doi.org/10.1038/s41392-023-01686-z
- Primary Citation of Related Structures:
8HXQ, 8HXR - PubMed Abstract:
Ligand-induced receptor dimerization or oligomerization is a widespread mechanism for ensuring communication specificity, safeguarding receptor activation, and facilitating amplification of signal transduction across the cellular membrane. However, cell-surface antigen-induced multimerization (dubbed AIM herein) has not yet been consciously leveraged in chimeric antigen receptor (CAR) engineering for enriching T cell-based therapies. We co-developed ciltacabtagene autoleucel (cilta-cel), whose CAR incorporates two B-cell maturation antigen (BCMA)-targeted nanobodies in tandem, for treating multiple myeloma. Here we elucidated a structural and functional model in which BCMA-induced cilta-cel CAR multimerization amplifies myeloma-targeted T cell-mediated cytotoxicity. Crystallographic analysis of BCMA-nanobody complexes revealed atomic details of antigen-antibody hetero-multimerization whilst analytical ultracentrifugation and small-angle X-ray scattering characterized interdependent BCMA apposition and CAR juxtaposition in solution. BCMA-induced nanobody CAR multimerization enhanced cytotoxicity, alongside elevated immune synapse formation and cytotoxicity-mediating cytokine release, towards myeloma-derived cells. Our results provide a framework for contemplating the AIM approach in designing next-generation CARs.
- Shanghai Institute of Hematology, National Research Center for Translational Medicine, State Key Laboratory of Medical Genomics, Ruijin Hospital Affiliated to Shanghai Jiao Tong University School of Medicine, Shanghai, 200025, China.
Organizational Affiliation: