A simple goniometer-compatible flow cell for serial synchrotron X-ray crystallography.
Ghosh, S., Zoric, D., Dahl, P., Bjelcic, M., Johannesson, J., Sandelin, E., Borjesson, P., Bjorling, A., Banacore, A., Edlund, P., Aurelius, O., Milas, M., Nan, J., Shilova, A., Gonzalez, A., Mueller, U., Branden, G., Neutze, R.(2023) J Appl Crystallogr 56: 449-460
- PubMed: 37032973
- DOI: https://doi.org/10.1107/S1600576723001036
- Primary Citation of Related Structures:
8HUA - PubMed Abstract:
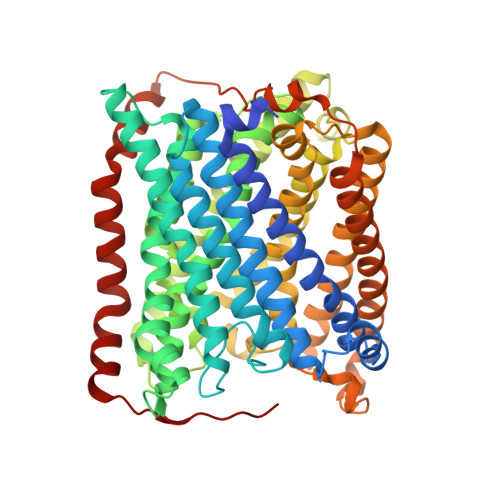
Serial femtosecond crystallography was initially developed for room-temperature X-ray diffraction studies of macromolecules at X-ray free electron lasers. When combined with tools that initiate biological reactions within microcrystals, time-resolved serial crystallography allows the study of structural changes that occur during an enzyme catalytic reaction. Serial synchrotron X-ray crystallography (SSX), which extends serial crystallography methods to synchrotron radiation sources, is expanding the scientific community using serial diffraction methods. This report presents a simple flow cell that can be used to deliver microcrystals across an X-ray beam during SSX studies. This device consists of an X-ray transparent glass capillary mounted on a goniometer-compatible 3D-printed support and is connected to a syringe pump via light-weight tubing. This flow cell is easily mounted and aligned, and it is disposable so can be rapidly replaced when blocked. This system was demonstrated by collecting SSX data at MAX IV Laboratory from microcrystals of the integral membrane protein cytochrome c oxidase from Thermus thermophilus , from which an X-ray structure was determined to 2.12 Å resolution. This simple SSX platform may help to lower entry barriers for non-expert users of SSX.
- Department of Chemistry and Molecular Biology, University of Gothenburg, Medicinaregatan 9C, 40530 Gothenburg, Sweden.
Organizational Affiliation: