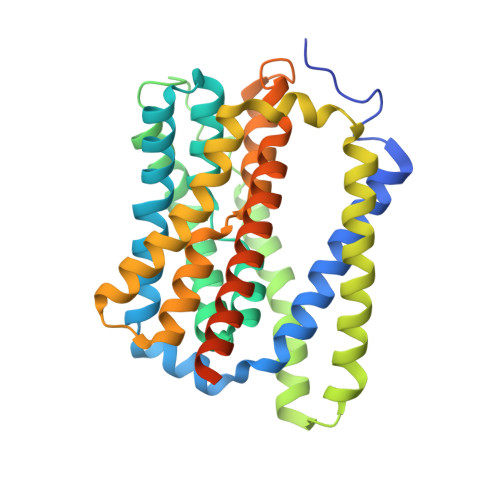
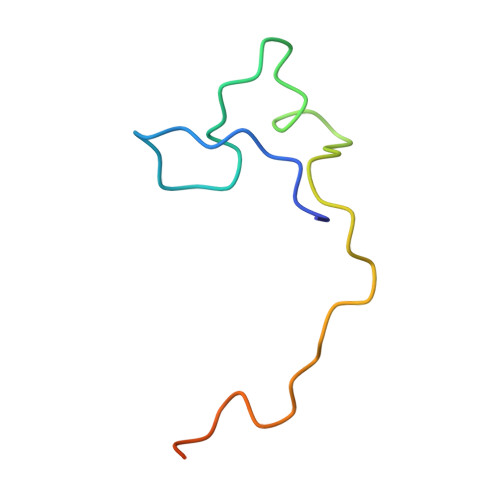
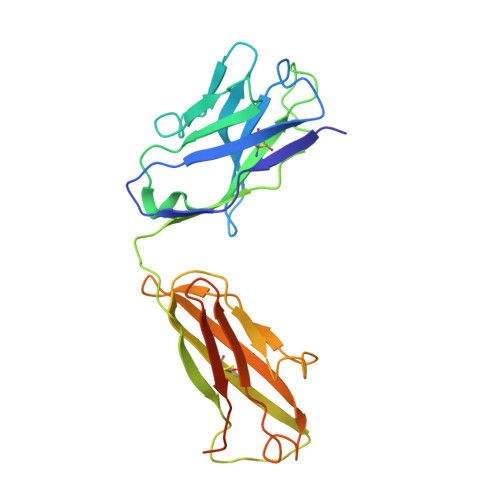
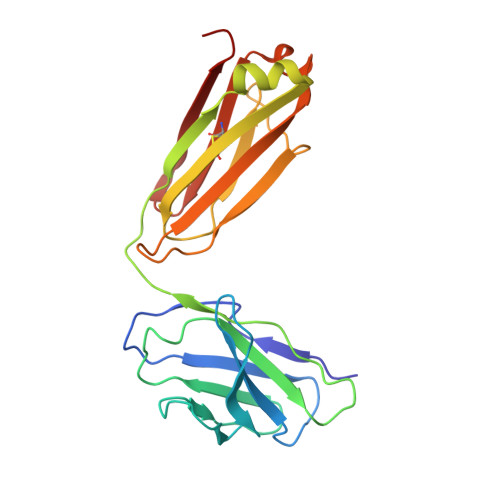
Structural basis of hepatitis B virus receptor binding.
Asami, J., Park, J.H., Nomura, Y., Kobayashi, C., Mifune, J., Ishimoto, N., Uemura, T., Liu, K., Sato, Y., Zhang, Z., Muramatsu, M., Wakita, T., Drew, D., Iwata, S., Shimizu, T., Watashi, K., Park, S.Y., Nomura, N., Ohto, U.(2024) Nat Struct Mol Biol 31: 447-454
- PubMed: 38233573
- DOI: https://doi.org/10.1038/s41594-023-01191-5
- Primary Citation of Related Structures:
8HRX, 8HRY - PubMed Abstract:
Hepatitis B virus (HBV), a leading cause of developing hepatocellular carcinoma affecting more than 290 million people worldwide, is an enveloped DNA virus specifically infecting hepatocytes. Myristoylated preS1 domain of the HBV large surface protein binds to the host receptor sodium-taurocholate cotransporting polypeptide (NTCP), a hepatocellular bile acid transporter, to initiate viral entry. Here, we report the cryogenic-electron microscopy structure of the myristoylated preS1 (residues 2-48) peptide bound to human NTCP. The unexpectedly folded N-terminal half of the peptide embeds deeply into the outward-facing tunnel of NTCP, whereas the C-terminal half formed extensive contacts on the extracellular surface. Our findings reveal an unprecedented induced-fit mechanism for establishing high-affinity virus-host attachment and provide a blueprint for the rational design of anti-HBV drugs targeting virus entry.
- Graduate School of Pharmaceutical Sciences, The University of Tokyo, Hongo, Bunkyo-ku, Tokyo, Japan.
Organizational Affiliation: