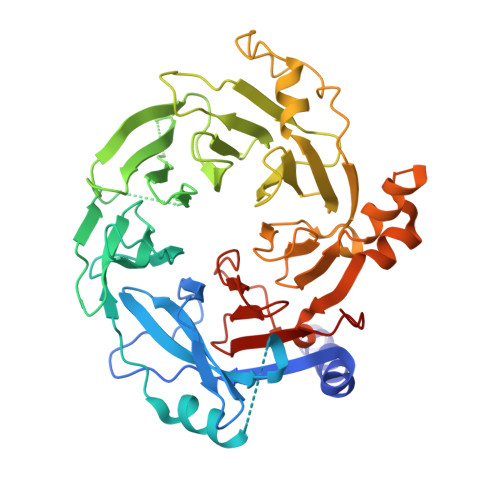
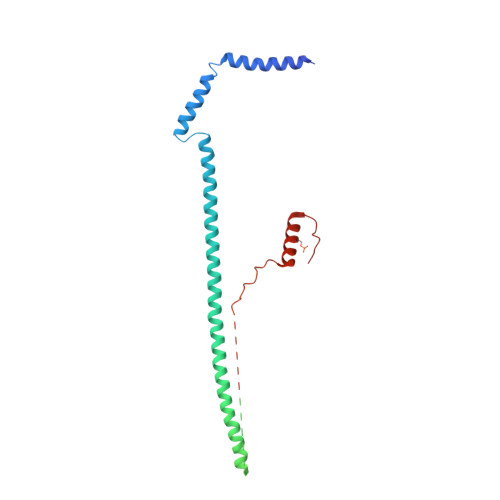
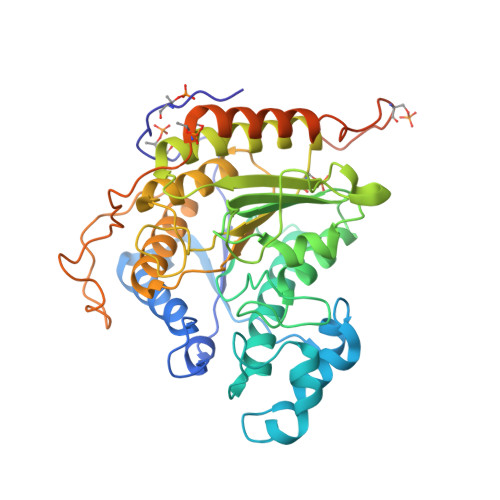
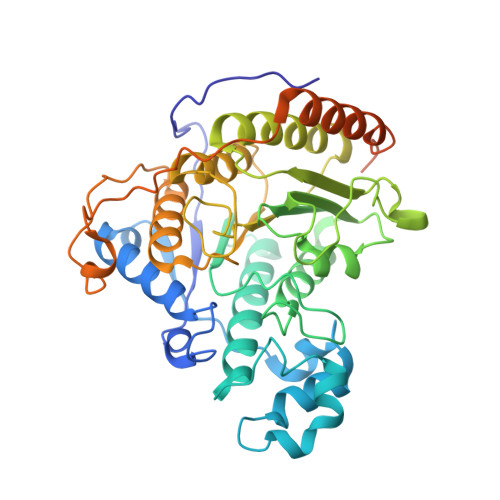
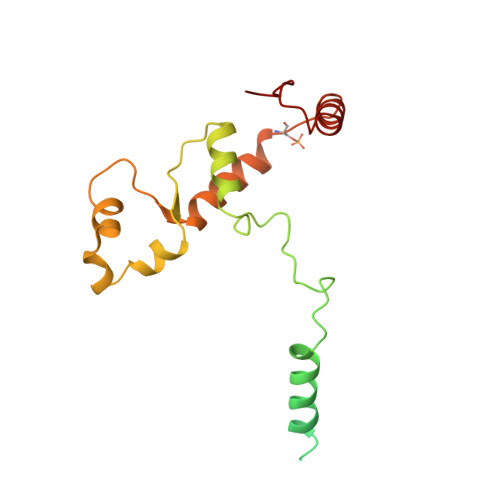
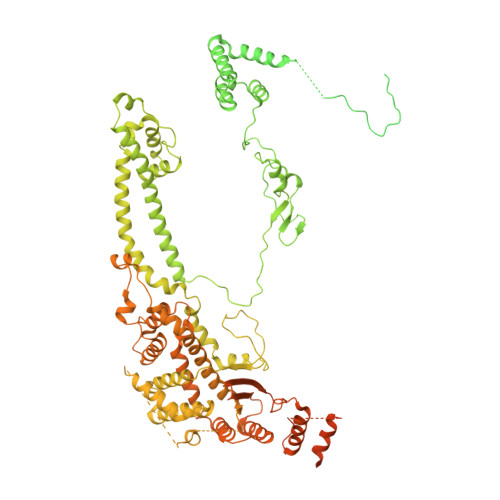
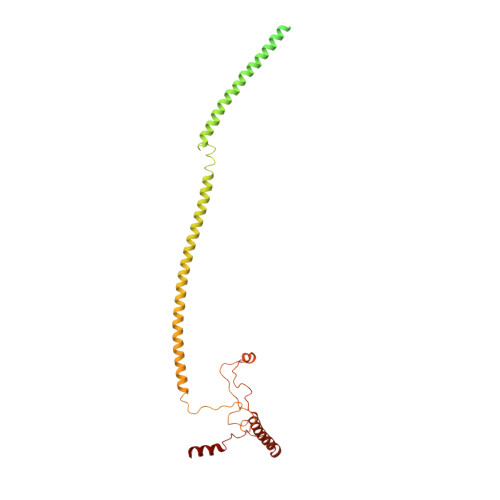
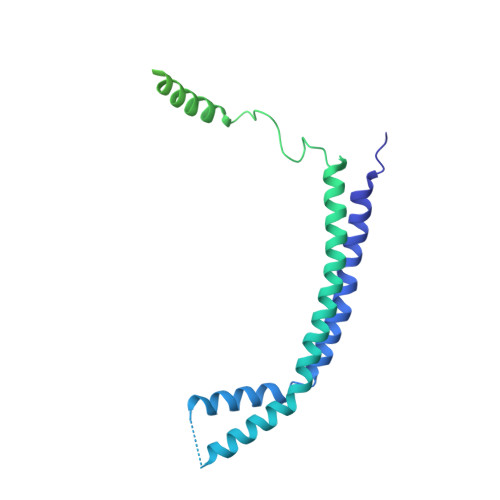
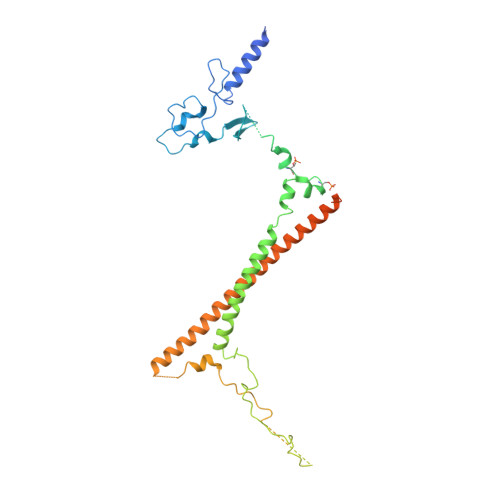
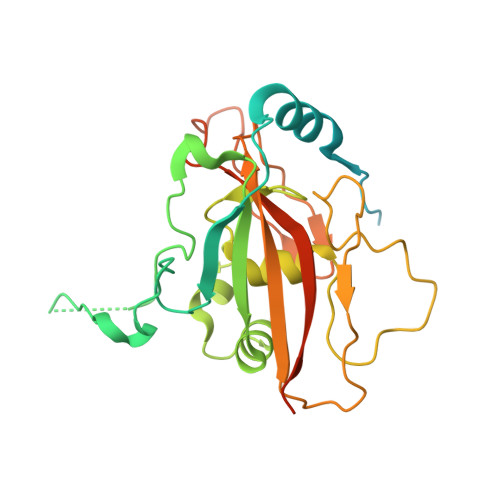
Structure of a SIN3-HDAC complex from budding yeast.
Guo, Z., Chu, C., Lu, Y., Zhang, X., Xiao, Y., Wu, M., Gao, S., Wong, C.C.L., Zhan, X., Wang, C.(2023) Nat Struct Mol Biol 30: 753-760
- PubMed: 37081318
- DOI: https://doi.org/10.1038/s41594-023-00975-z
- Primary Citation of Related Structures:
8HPO - PubMed Abstract:
SIN3-HDAC (histone deacetylases) complexes have important roles in facilitating local histone deacetylation to regulate chromatin accessibility and gene expression. Here, we present the cryo-EM structure of the budding yeast SIN3-HDAC complex Rpd3L at an average resolution of 2.6 Å. The structure reveals that two distinct arms (ARM1 and ARM2) hang on a T-shaped scaffold formed by two coiled-coil domains. In each arm, Sin3 interacts with different subunits to create a different environment for the histone deacetylase Rpd3. ARM1 is in the inhibited state with the active site of Rpd3 blocked, whereas ARM2 is in an open conformation with the active site of Rpd3 exposed to the exterior space. The observed asymmetric architecture of Rpd3L is different from those of available structures of other class I HDAC complexes. Our study reveals the organization mechanism of the SIN3-HDAC complex and provides insights into the interaction pattern by which it targets histone deacetylase to chromatin.
- College of Life Sciences, Zhejiang University, Hangzhou, China.
Organizational Affiliation: