Production of D-p-hydroxyphenylglycine by double-enzyme cascade
Zhang, L.D., Song, W.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| N-carbamoyl-D-amino acid hydrolase | 304 | Agrobacterium sp. KNK712 | Mutation(s): 1 EC: 3.5.1.77 | 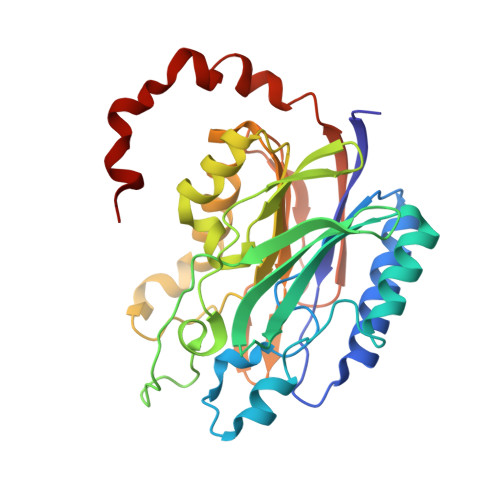 | |
UniProt | |||||
Find proteins for P60327 (Agrobacterium sp. (strain KNK712)) Explore P60327 Go to UniProtKB: P60327 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P60327 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| M9L (Subject of Investigation/LOI) Query on M9L | I [auth A] J [auth B] K [auth C] L [auth D] M [auth E] | (2~{R})-2-(aminocarbonylamino)-2-(4-hydroxyphenyl)ethanoic acid C9 H10 N2 O4 GSHIDXLOTQDUAV-SSDOTTSWSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 79.165 | α = 91.4 |
| b = 79.865 | β = 98.48 |
| c = 91.07 | γ = 92.78 |
| Software Name | Purpose |
|---|---|
| Aimless | data scaling |
| REFMAC | refinement |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |