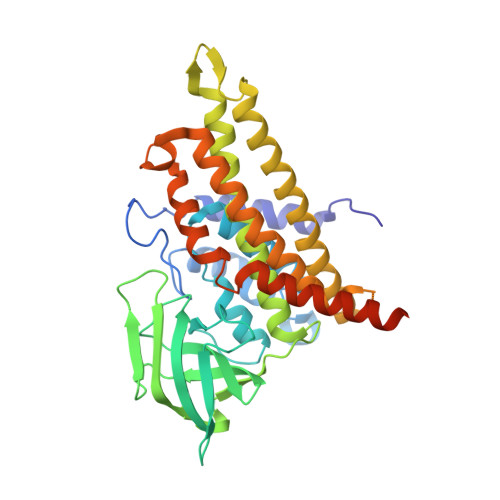
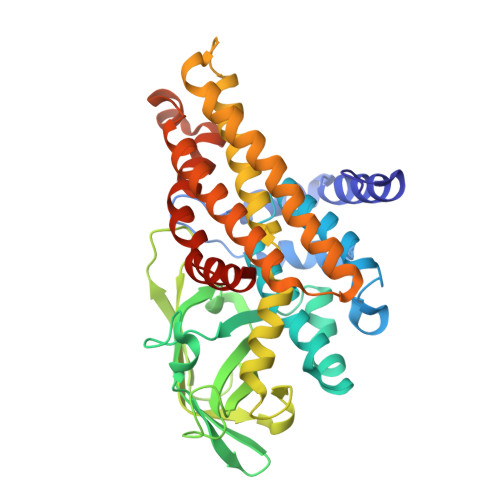
Oxidase Heterotetramer Completes 1-Azabicyclo[3.1.0]hexane Formation with the Association of a Nonribosomal Peptide Synthetase.
Cheng, Y., Yi, X., Zhang, Y., He, Q., Chen, D., Cao, W., Fang, P., Liu, W.(2023) J Am Chem Soc 145: 8896-8907
- PubMed: 37043819
- DOI: https://doi.org/10.1021/jacs.2c12507
- Primary Citation of Related Structures:
8GS1, 8HK0 - PubMed Abstract:
Ficellomycin, azinomycins, and vazabitide A are nonribosomal peptide natural products characterized by an amino acid unit that contains a similar 1- a za b i c yclo[3.1.0] h exane (ABCH) pharmacophore. This unit is derived from d i a mino- d ihydroxy- h eptanic acid (DADH); however, the process through which linear DADH is cyclized to furnish an ABCH ring system remains poorly understood. Based on the reconstitution of the route of the ABCH-containing unit by blending genes/enzymes involved in the biosynthesis of ficellomycin and azinomycins, we report that ABCH formation is completed by an oxidase heterotetramer with the association of a nonribosomal peptide synthetase (NRPS). The DADH precursor was prepared in Escherichia coli to produce a conjugate subjected to in vitro enzymatic hydrolysis for offloading from an amino-group carrier protein. To furnish an aziridine ring, DADH was processed by C7-hydroxyl sulfonation and sulfate elimination-coupled cyclization. Further cyclization leading to an azabicyclic hexane pharmacophore was proved to occur in the NRPS, where the oxidase heterotetramer functions in trans and catalyzes α,β-dehydrogenation to initiate the formation of a fused five-membered nitrogen heterocycle. The identity of ABCH was validated by utilization of the resultant ABCH-containing unit in the total biosynthesis of ficellomycin. Biochemical characterization, crystal structure, and site-specific mutagenesis rationalize the catalytic mechanism of the unusual oxidase heterotetramer.
- State Key Laboratory of Bioorganic and Natural Products Chemistry, Center for Excellence in Molecular Synthesis, Shanghai Institute of Organic Chemistry, University of Chinese Academy of Sciences, 345 Lingling Road, Shanghai 200032, China.
Organizational Affiliation: