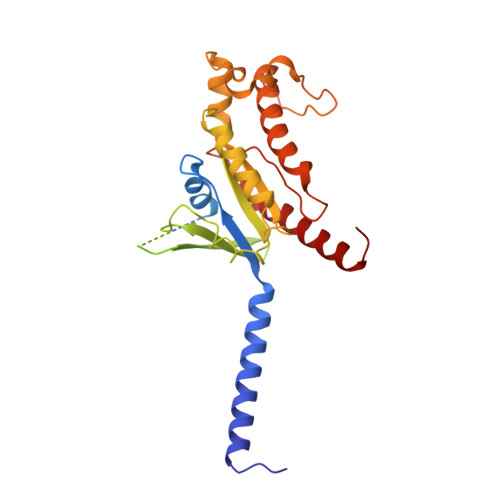
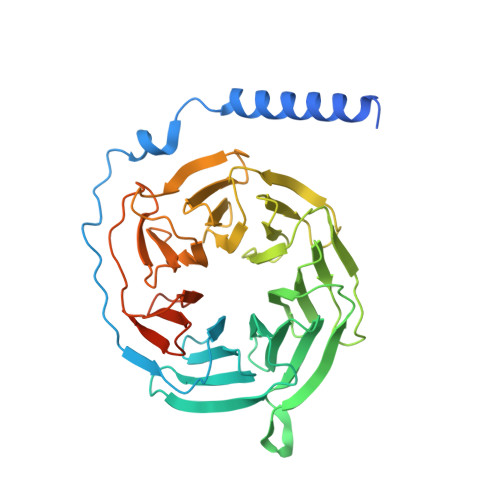
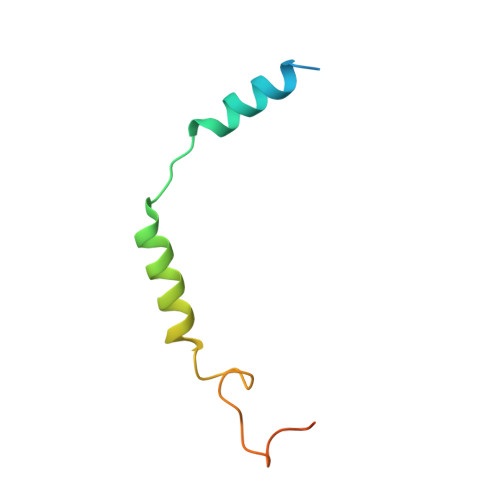
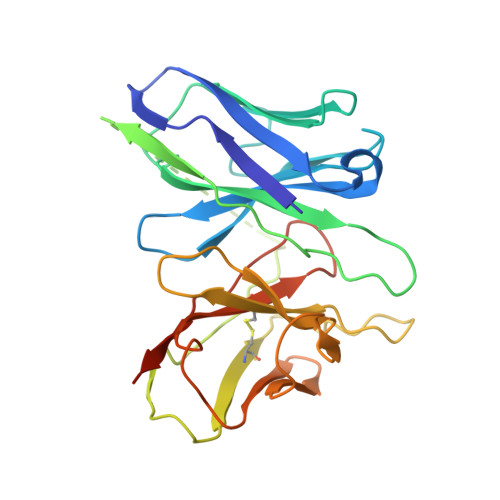
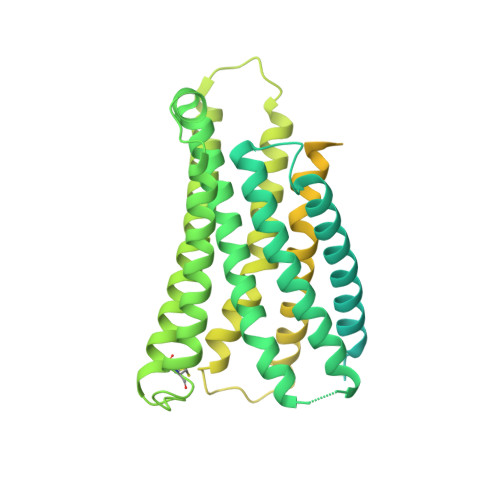
Mechanism of agonist-induced activation of the human itch receptor MRGPRX1.
Gan, B., Yu, L., Yang, H., Jiao, H., Pang, B., Chen, Y., Wang, C., Lv, R., Hu, H., Cao, Z., Ren, R.(2023) PLoS Biol 21: e3001975-e3001975
- PubMed: 37347749
- DOI: https://doi.org/10.1371/journal.pbio.3001975
- Primary Citation of Related Structures:
8HJ5 - PubMed Abstract:
Mas-related G-protein-coupled receptors X1-X4 (MRGPRX1-X4) are 4 primate-specific receptors that are recently reported to be responsible for many biological processes, including itch sensation, pain transmission, and inflammatory reactions. MRGPRX1 is the first identified human MRGPR, and its expression is restricted to primary sensory neurons. Due to its dual roles in itch and pain signaling pathways, MRGPRX1 has been regarded as a promising target for itch remission and pain inhibition. Here, we reported a cryo-electron microscopy (cryo-EM) structure of Gq-coupled MRGPRX1 in complex with a synthetic agonist compound 16 in an active conformation at an overall resolution of 3.0 Å via a NanoBiT tethering strategy. Compound 16 is a new pain-relieving compound with high potency and selectivity to MRGPRX1 over other MRGPRXs and opioid receptor. MRGPRX1 was revealed to share common structural features of the Gq-mediated receptor activation mechanism of MRGPRX family members, but the variable residues in orthosteric pocket of MRGPRX1 exhibit the unique agonist recognition pattern, potentially facilitating to design MRGPRX1-specific modulators. Together with receptor activation and itch behavior evaluation assays, our study provides a structural snapshot to modify therapeutic molecules for itch relieving and analgesia targeting MRGPRX1.
- Shanghai Key Laboratory of Metabolic Remodeling and Health, Institute of Metabolism and Integrative Biology, Fudan University, Shanghai, China.
Organizational Affiliation: