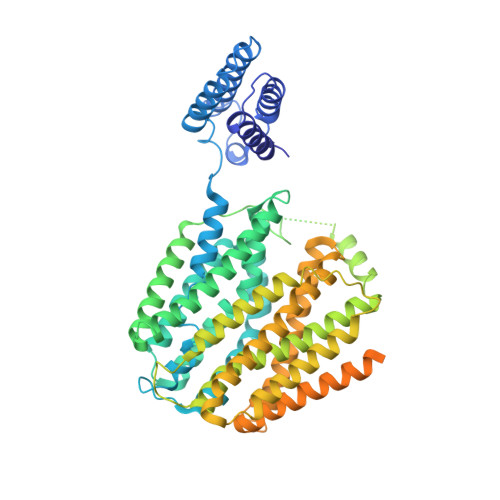
Molecular mechanism of substrate recognition by folate transporter SLC19A1.
Dang, Y., Zhou, D., Du, X., Zhao, H., Lee, C.H., Yang, J., Wang, Y., Qin, C., Guo, Z., Zhang, Z.(2022) Cell Discov 8: 141-141
- PubMed: 36575193
- DOI: https://doi.org/10.1038/s41421-022-00508-w
- Primary Citation of Related Structures:
8HII, 8HIJ, 8HIK - PubMed Abstract:
Folate (vitamin B 9 ) is the coenzyme involved in one-carbon transfer biochemical reactions essential for cell survival and proliferation, with its inadequacy causing developmental defects or severe diseases. Notably, mammalian cells lack the ability to de novo synthesize folate but instead rely on its intake from extracellular sources via specific transporters or receptors, among which SLC19A1 is the ubiquitously expressed one in tissues. However, the mechanism of substrate recognition by SLC19A1 remains unclear. Here we report the cryo-EM structures of human SLC19A1 and its complex with 5-methyltetrahydrofolate at 3.5-3.6 Å resolution and elucidate the critical residues for substrate recognition. In particular, we reveal that two variant residues among SLC19 subfamily members designate the specificity for folate. Moreover, we identify intracellular thiamine pyrophosphate as the favorite coupled substrate for folate transport by SLC19A1. Together, this work establishes the molecular basis of substrate recognition by this central folate transporter.
- State Key Laboratory of Membrane Biology, Peking University-Tsinghua University-National Institute of Biological Sciences Joint Graduate Program, Academy for Advanced Interdisciplinary Studies, Peking University, Beijing, China.
Organizational Affiliation: