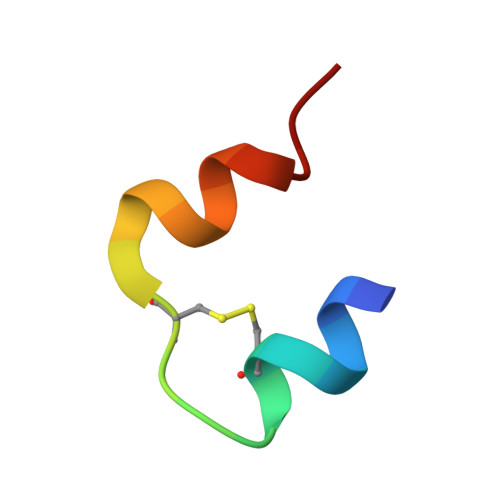
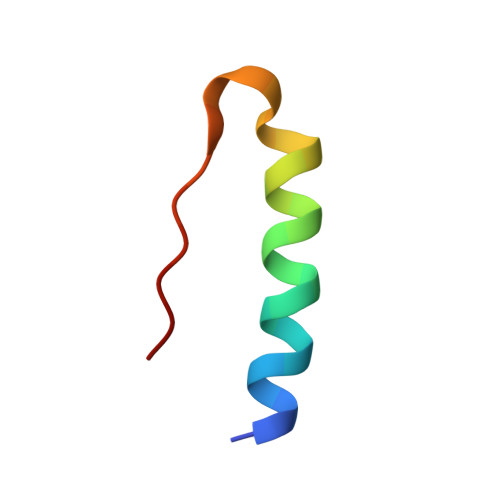
Comparative Study of High-Resolution LysB29(N epsilon-myristoyl) des(B30) Insulin Structures Display Novel Dynamic Causal Interrelations in Monomeric-Dimeric Motions
Ayan, E., Destan, E., Kepceoglu, A., Ciftci, H.I., Kati, A., DeMirci, H.(2023) Crystals (Basel) 13