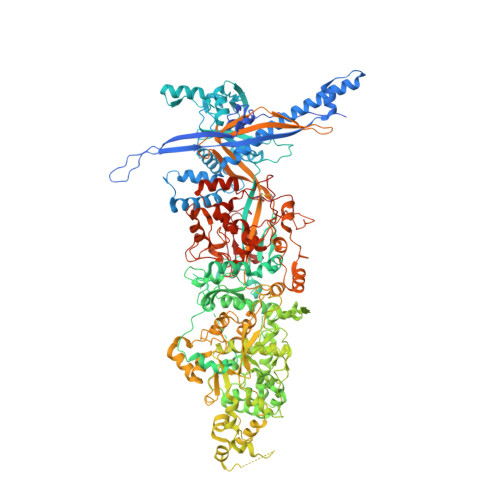
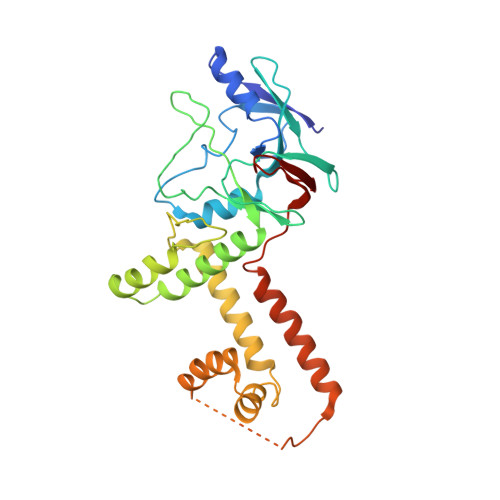
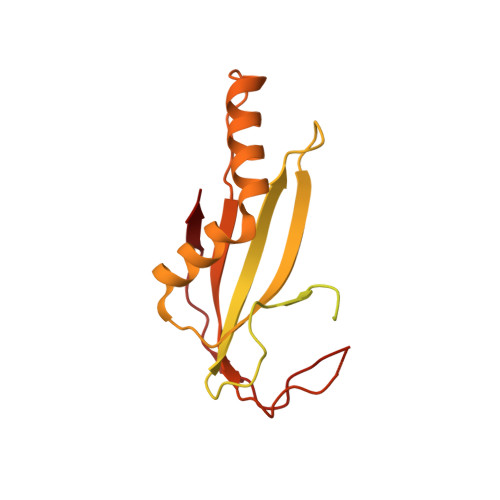
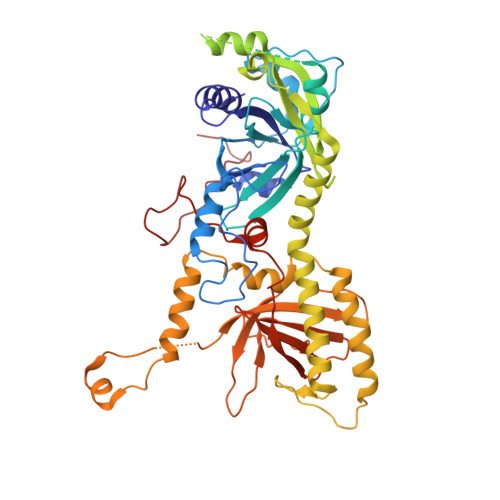
Cryo-electron microscopy structures of capsids and in situ portals of DNA-devoid capsids of human cytomegalovirus.
Li, Z., Pang, J., Gao, R., Wang, Q., Zhang, M., Yu, X.(2023) Nat Commun 14: 2025-2025
- PubMed: 37041152
- DOI: https://doi.org/10.1038/s41467-023-37779-0
- Primary Citation of Related Structures:
8HEU, 8HEV, 8HEX, 8HEY - PubMed Abstract:
The portal-scaffold complex is believed to nucleate the assembly of herpesvirus procapsids. During capsid maturation, two events occur: scaffold expulsion and DNA incorporation. The portal-scaffold interaction and the conformational changes that occur to the portal during the different stages of capsid formation have yet to be elucidated structurally. Here we present high-resolution structures of the A- and B-capsids and in-situ portals of human cytomegalovirus. We show that scaffolds bind to the hydrophobic cavities formed by the dimerization and Johnson-fold domains of the major capsid proteins. We further show that 12 loop-helix-loop fragments-presumably from the scaffold domain-insert into the hydrophobic pocket of the portal crown domain. The portal also undergoes significant changes both positionally and conformationally as it accompanies DNA packaging. These findings unravel the mechanism by which the portal interacts with the scaffold to nucleate capsid assembly and further our understanding of scaffold expulsion and DNA incorporation.
- School of Pharmaceutical Science and Technology, Hangzhou Institute for Advanced Study, University of Chinese Academy of Sciences, Hangzhou, 310024, China.
Organizational Affiliation: