Structural insight into the septal peptidoglycan hydrolysis machinery of bacterial cell division
Zhang, Z., Dong, H., Chen, Y.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cell division ATP-binding protein FtsE | 222 | Escherichia coli K-12 | Mutation(s): 1 Gene Names: ftsE, b3463, JW3428 Membrane Entity: Yes | 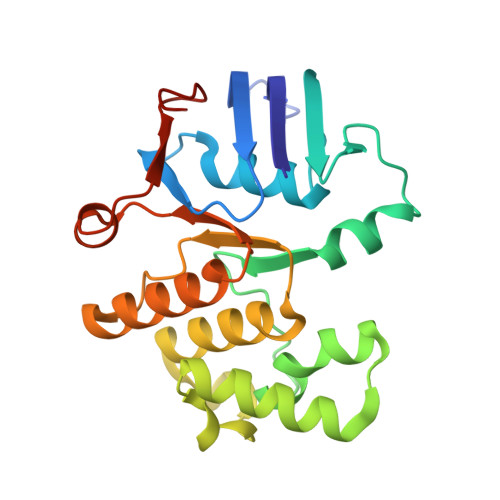 | |
UniProt | |||||
Find proteins for P0A9R7 (Escherichia coli (strain K12)) Explore P0A9R7 Go to UniProtKB: P0A9R7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A9R7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cell division protein FtsX | 352 | Escherichia coli K-12 | Mutation(s): 0 Gene Names: ftsX, ftsS, b3462, JW3427 Membrane Entity: Yes | 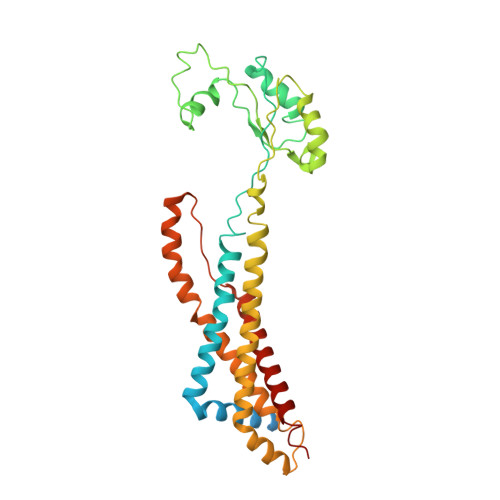 | |
UniProt | |||||
Find proteins for P0AC30 (Escherichia coli (strain K12)) Explore P0AC30 Go to UniProtKB: P0AC30 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0AC30 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Murein hydrolase activator EnvC | 419 | Escherichia coli K-12 | Mutation(s): 0 Gene Names: envC, yibP, b3613, JW5646 Membrane Entity: Yes | 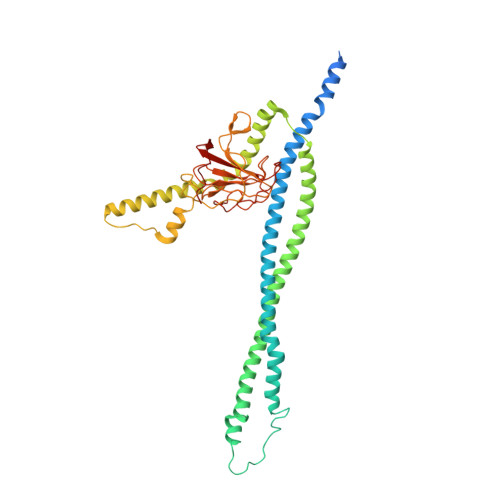 | |
UniProt | |||||
Find proteins for P37690 (Escherichia coli (strain K12)) Explore P37690 Go to UniProtKB: P37690 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P37690 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ATP (Subject of Investigation/LOI) Query on ATP | F [auth A], G [auth B] | ADENOSINE-5'-TRIPHOSPHATE C10 H16 N5 O13 P3 ZKHQWZAMYRWXGA-KQYNXXCUSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| RECONSTRUCTION | cryoSPARC |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | 31800052 |
| Ministry of Science and Technology (MoST, China) | China | 2018YFA093200 |