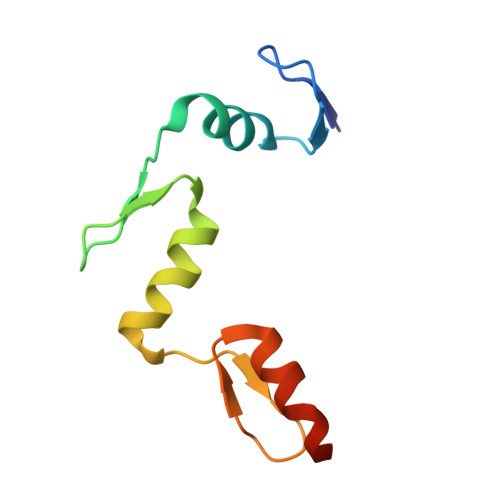
ZBTB7A regulates primed-to-naive transition of pluripotent stem cells via recognition of the PNT-associated sequence by zinc fingers 1-2 and recognition of gamma-globin -200 gene element by zinc fingers 1-4.
Yang, Y., Xiao, L., Xue, Y., Idris, M.O., Liu, J., Pei, D., Shi, Y., Liao, B., Li, F.(2023) FEBS J 290: 3896-3909
- PubMed: 37013936
- DOI: https://doi.org/10.1111/febs.16789
- Primary Citation of Related Structures:
8H9H - PubMed Abstract:
ZBTB7A, a transcription factor containing a tandem array of four Cys2-His2 zinc fingers (ZFs), is vital for multiple physiological events through directional binding to different genomic loci. Our previously determined crystal structure of ZBTB7A in complex with a GCCCCTTCCCC sequence revealed that all four ZFs (ZF1-4) are involved in binding to γ-globin -200 gene element to repress fetal haemoglobin expression. Recently, it has been reported that ZBTB7A drives primed-to-naïve transition (PNT) of pluripotent stem cells through binding to a 12-bp consensus sequence ([AAGGACCCAGAT], referred to as PNT-associated sequence). Here, we report a crystal structure of ZBTB7A ZF1-3 in complex with the PNT-associated sequence. The structure shows that ZF1 and ZF2 primarily contribute to recognizing the GACCC core sequence mimicking the half part (GCCCC) of γ-globin -200 gene element via specific hydrogen bonding and van der Waals contacts. The mutations of key residues in ZF1-2 remarkably reduce their binding affinities for the PNT-associated sequence in vitro and cannot restore epiblast stem cells to the naïve pluripotent state in vivo. Collectively, our studies demonstrate that ZBTB7A mainly employs its ZF1-2 to recognize the PNT-associated sequence but recognizes γ-globin -200 gene element via ZF1-4, providing insights into the molecular mechanism for the diversity of ZBTB7A's genomic localization.
- Hefei National Research Center for Cross Disciplinary Science, Division of Life Sciences and Medicine, School of Life Sciences, University of Science and Technology of China, Hefei, China.
Organizational Affiliation: