A Capsid Structure of Ralstonia solanacearum podoviridae GP4 with a Triangulation Number T = 9.
Zheng, J., Chen, W., Xiao, H., Yang, F., Li, X., Song, J., Cheng, L., Liu, H.(2022) Viruses 14
- PubMed: 36366529
- DOI: https://doi.org/10.3390/v14112431
- Primary Citation of Related Structures:
8H89 - PubMed Abstract:
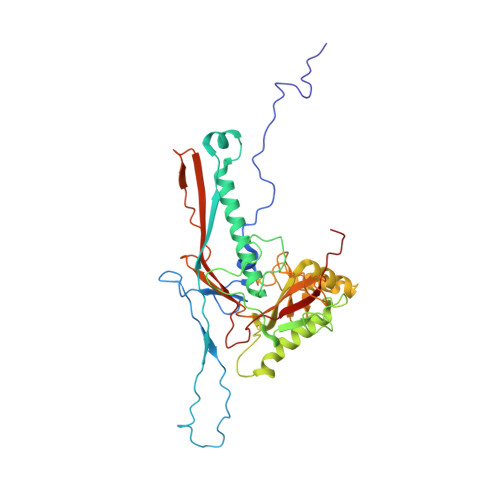
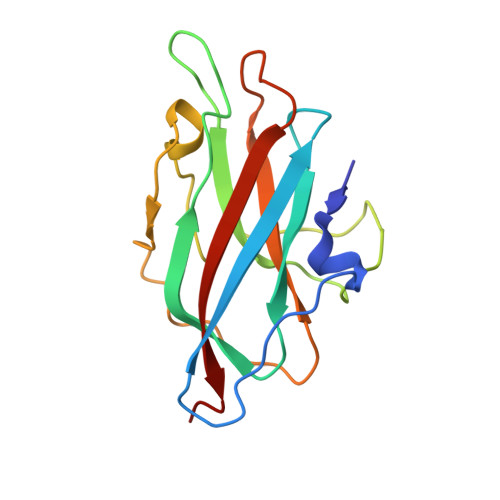
GP4, a new Ralstonia solanacearum phage, is a short-tailed phage. Few structures of Ralstonia solanacearum phages have been resolved to near-atomic resolution until now. Here, we present a 3.7 Å resolution structure of the GP4 head by cryo-electron microscopy (cryo-EM). The GP4 head contains 540 copies of major capsid protein (MCP) gp2 and 540 copies of cement protein (CP) gp1 arranged in an icosahedral shell with a triangulation number T = 9. The structures of gp2 and gp1 show a canonical HK97-like fold and an Ig-like fold, respectively. The trimeric CPs stick on the surface of the head along the quasi-threefold axis of the icosahedron generating a sandwiched three-layer electrostatic complementary potential, thereby enhancing the head stability. The assembly pattern of the GP4 head provides a platform for the further exploration of the interaction between Ralstonia solanacearum and corresponding phages.
- Institute of Interdisciplinary Studies, Key Laboratory for Matter Microstructure and Function of Hunan Province, Key Laboratory of Low-Dimensional Quantum Structures and Quantum Control, Hunan Normal University, Changsha 410082, China.
Organizational Affiliation: