Crystal structure of aqualigase bound with Suc-AAPF
Li, H., Ma, M.Z., Zhang, L.J., Dai, L., Chen, C.-C., Guo, R.-T.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Subtilisin | 281 | Bacillus subtilis | Mutation(s): 0 Gene Names: KS08_04965 EC: 3.4.21.62 | 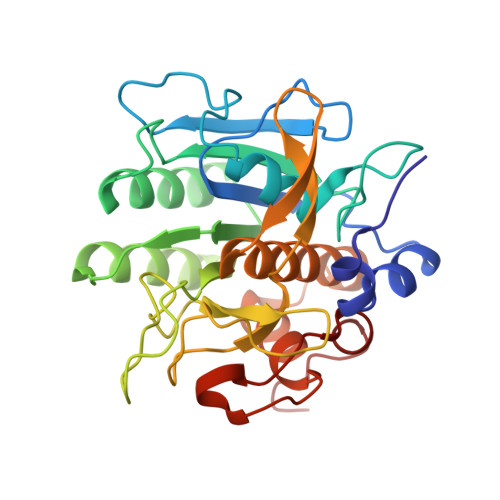 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 5,6-DIHYDRO-BENZO[H]CINNOLIN-3-YLAMINE | 6 | synthetic construct | Mutation(s): 0 |  | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PGE Query on PGE | D [auth A] | TRIETHYLENE GLYCOL C6 H14 O4 ZIBGPFATKBEMQZ-UHFFFAOYSA-N |  | ||
| CA Query on CA | C [auth A] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 36.578 | α = 90 |
| b = 71.68 | β = 114.74 |
| c = 40.804 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| SAINT | data scaling |
| PDB_EXTRACT | data extraction |
| SAINT | data reduction |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |