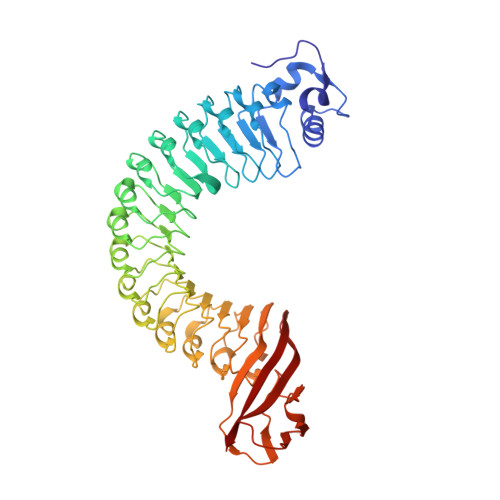
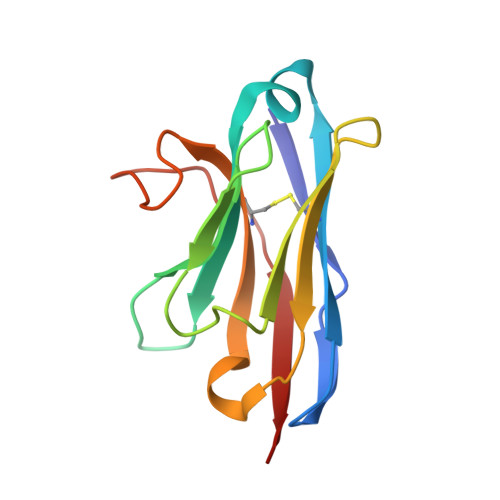
Anti-InlA single-domain antibodies that inhibit the cell invasion of Listeria monocytogenes.
Yamazaki, T., Nagatoishi, S., Yamawaki, T., Nozawa, T., Matsunaga, R., Nakakido, M., Caaveiro, J.M.M., Nakagawa, I., Tsumoto, K.(2023) J Biological Chem 299: 105254-105254
- PubMed: 37716701
- DOI: https://doi.org/10.1016/j.jbc.2023.105254
- Primary Citation of Related Structures:
8H62, 8H63, 8H64 - PubMed Abstract:
Listeriosis, caused by infection with Listeria monocytogenes, is a severe disease with a high mortality rate. The L. monocytogenes virulence factor, internalin family protein InlA, which binds to the host receptor E-cadherin, is necessary to invade host cells. Here, we isolated two single-domain antibodies (V H Hs) that bind to InlA with picomolar affinities from an alpaca immune library using the phage display method. These InlA-specific V H Hs inhibited the binding of InlA to the extracellular domains of E-cadherin in vitro as shown by biophysical interaction analysis. Furthermore, we determined that the V H Hs inhibited the invasion of L. monocytogenes into host cells in culture. High-resolution X-ray structure analyses of the complexes of V H Hs with InlA revealed that the V H Hs bind to the same binding site as E-cadherin against InlA. We conclude that these V H Hs have the potential for use as drugs to treat listeriosis.
- Department of Bioengineering, School of Engineering, The University of Tokyo, Tokyo, Japan.
Organizational Affiliation: