A case of balance engineering exhibits kinetic relationship between mesophilic and thermophilic poly(ethylene terephthalate) depolymerases.
Lee, S.H., Seo, H., Kim, K.J.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Poly(ethylene terephthalate) hydrolase | 300 | Piscinibacter sakaiensis | Mutation(s): 8 Gene Names: ISF6_4831 EC: 3.1.1.101 | 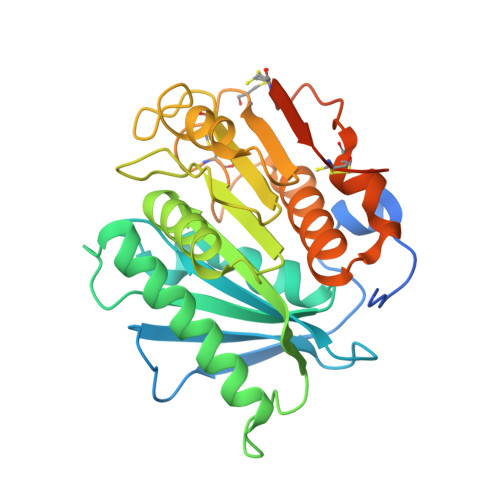 | |
UniProt | |||||
Find proteins for A0A0K8P6T7 (Piscinibacter sakaiensis) Explore A0A0K8P6T7 Go to UniProtKB: A0A0K8P6T7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A0K8P6T7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PO4 Query on PO4 | D [auth A], E [auth A], G [auth B] | PHOSPHATE ION O4 P NBIIXXVUZAFLBC-UHFFFAOYSA-K |  | ||
| GOL Query on GOL | H [auth B] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| K Query on K | C [auth A], F [auth B], I [auth B] | POTASSIUM ION K NPYPAHLBTDXSSS-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 139.218 | α = 90 |
| b = 50.665 | β = 124.64 |
| c = 80.854 | γ = 90 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data reduction |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data scaling |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Other government | -- |