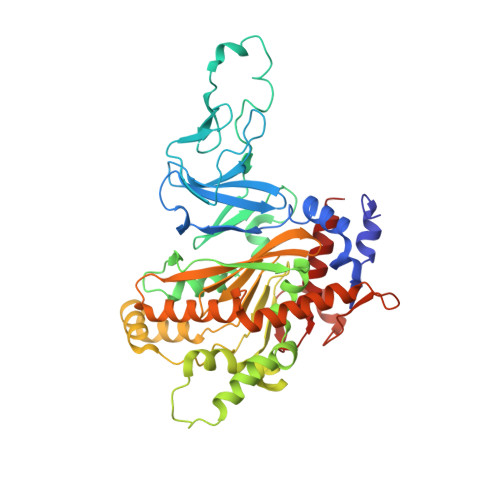
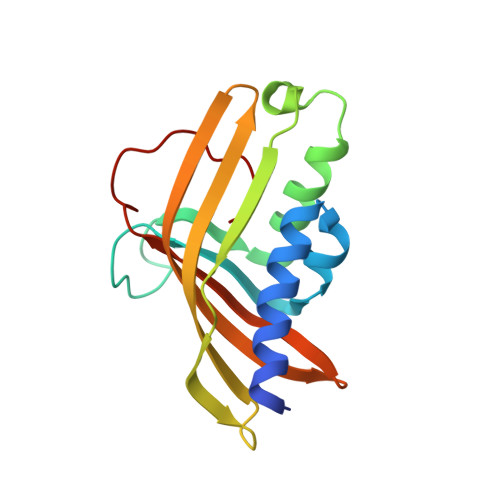
Structural and biochemical characterization of the key components of an auxin degradation operon from the rhizosphere bacterium Variovorax.
Ma, Y., Li, X., Wang, F., Zhang, L., Zhou, S., Che, X., Yu, D., Liu, X., Li, Z., Sun, H., Yu, G., Zhang, H.(2023) PLoS Biol 21: e3002189-e3002189
- PubMed: 37459330
- DOI: https://doi.org/10.1371/journal.pbio.3002189
- Primary Citation of Related Structures:
8H2T - PubMed Abstract:
Plant-associated bacteria play important regulatory roles in modulating plant hormone auxin levels, affecting the growth and yields of crops. A conserved auxin degradation (iad) operon was recently identified in the Variovorax genomes, which is responsible for root growth inhibition (RGI) reversion, promoting rhizosphere colonization and root growth. However, the molecular mechanism underlying auxin degradation by Variovorax remains unclear. Here, we systematically screened Variovorax iad operon products and identified 2 proteins, IadK2 and IadD, that directly associate with auxin indole-3-acetic acid (IAA). Further biochemical and structural studies revealed that IadK2 is a highly IAA-specific ATP-binding cassette (ABC) transporter solute-binding protein (SBP), likely involved in IAA uptake. IadD interacts with IadE to form a functional Rieske non-heme dioxygenase, which works in concert with a FMN-type reductase encoded by gene iadC to transform IAA into the biologically inactive 2-oxindole-3-acetic acid (oxIAA), representing a new bacterial pathway for IAA inactivation/degradation. Importantly, incorporation of a minimum set of iadC/D/E genes could enable IAA transformation by Escherichia coli, suggesting a promising strategy for repurposing the iad operon for IAA regulation. Together, our study identifies the key components and underlying mechanisms involved in IAA transformation by Variovorax and brings new insights into the bacterial turnover of plant hormones, which would provide the basis for potential applications in rhizosphere optimization and ecological agriculture.
- State Key Laboratory of Experimental Hematology, Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, Haihe Laboratory of Cell Ecosystem, Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University, Tianjin, China.
Organizational Affiliation: