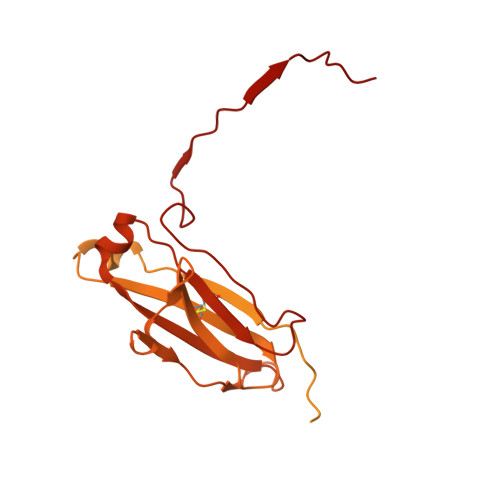
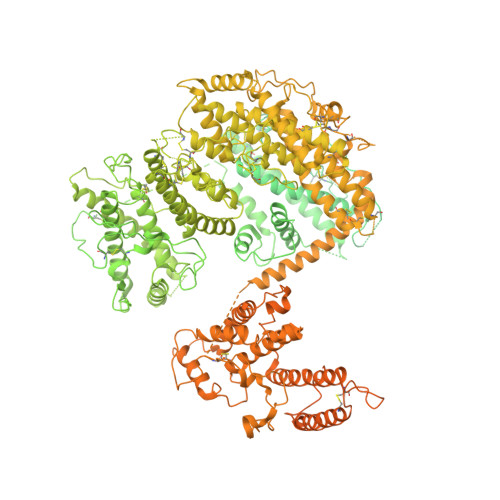
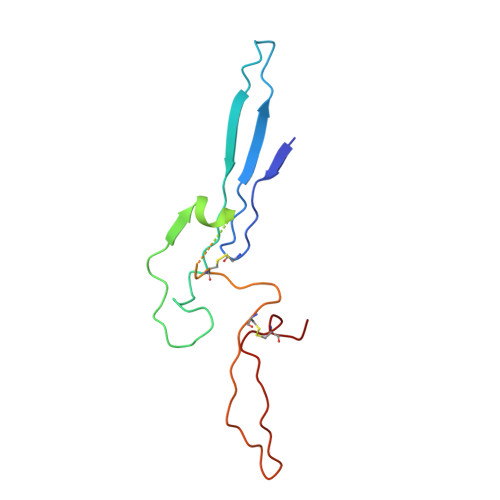
Cryo-electron microscopy of IgM-VAR2CSA complex reveals IgM inhibits binding of Plasmodium falciparum to Chondroitin Sulfate A.
Akhouri, R.R., Goel, S., Skoglund, U.(2023) Nat Commun 14: 6391-6391
- PubMed: 37828011
- DOI: https://doi.org/10.1038/s41467-023-41838-x
- Primary Citation of Related Structures:
8GZN - PubMed Abstract:
Placental malaria is caused by Plasmodium falciparum-infected erythrocytes (IEs) adhering to chondroitin sulfate proteoglycans in placenta via VAR2CSA-type PfEMP1. Human pentameric immunoglobulin M (IgM) binds to several types of PfEMP1, including VAR2CSA via its Fc domain. Here, a 3.6 Å cryo-electron microscopy map of the IgM-VAR2CSA complex reveals that two molecules of VAR2CSA bind to the Cµ4 of IgM through their DBL3X and DBL5ε domains. The clockwise and anti-clockwise rotation of the two VAR2CSA molecules on opposite faces of IgM juxtaposes C-termini of both VAR2CSA near the J chain, where IgM creates a wall between both VAR2CSA molecules and hinders its interaction with its receptor. To support this, we show when VAR2CSA is bound to IgM, its staining on IEs as well as binding of IEs to chondroitin sulfate A in vitro is severely compromised.
- Okinawa Institute of Science and Technology Graduate University, Okinawa, Japan. akhourirr@gmail.com.
Organizational Affiliation: