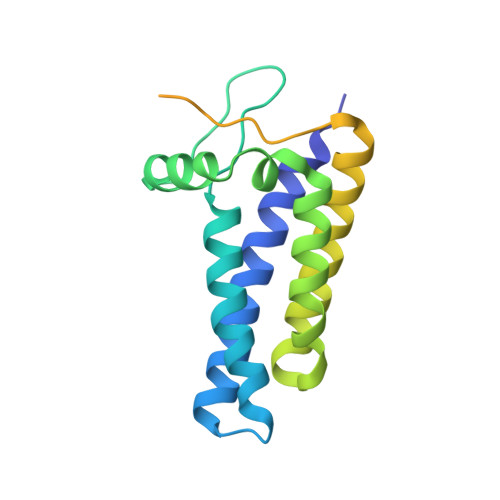
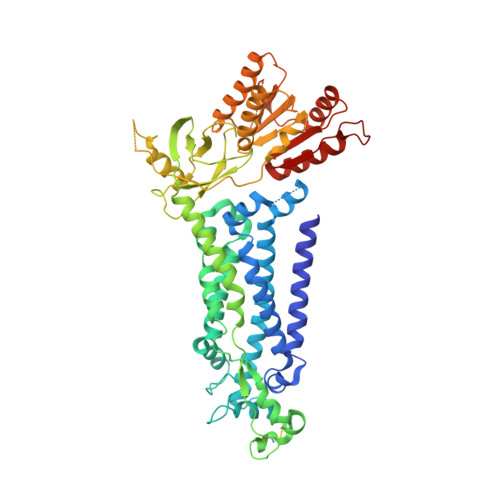
Structure of human phagocyte NADPH oxidase in the resting state.
Liu, R., Song, K., Wu, J.X., Geng, X.P., Zheng, L., Gao, X., Peng, H., Chen, L.(2022) Elife 11
- PubMed: 36413210
- DOI: https://doi.org/10.7554/eLife.83743
- Primary Citation of Related Structures:
8GZ3 - PubMed Abstract:
Phagocyte oxidase plays an essential role in the first line of host defense against pathogens. It oxidizes intracellular NADPH to reduce extracellular oxygen to produce superoxide anions that participate in pathogen killing. The resting phagocyte oxidase is a heterodimeric complex formed by two transmembrane proteins NOX2 and p22. Despite the physiological importance of this complex, its structure remains elusive. Here, we reported the cryo-EM structure of the functional human NOX2-p22 complex in nanodisc in the resting state. NOX2 shows a canonical 6-TM architecture of NOX and p22 has four transmembrane helices. M3, M4, and M5 of NOX2, and M1 and M4 helices of p22 are involved in the heterodimer formation. Dehydrogenase (DH) domain of NOX2 in the resting state is not optimally docked onto the transmembrane domain, leading to inefficient electron transfer and NADPH binding. Structural analysis suggests that the cytosolic factors might activate the NOX2-p22 complex by stabilizing the DH in a productive docked conformation.
- State Key Laboratory of Membrane Biology, College of Future Technology, Institute of Molecular Medicine, Peking University, Beijing Key Laboratory of Cardiometabolic Molecular Medicine, Beijing, China.
Organizational Affiliation: