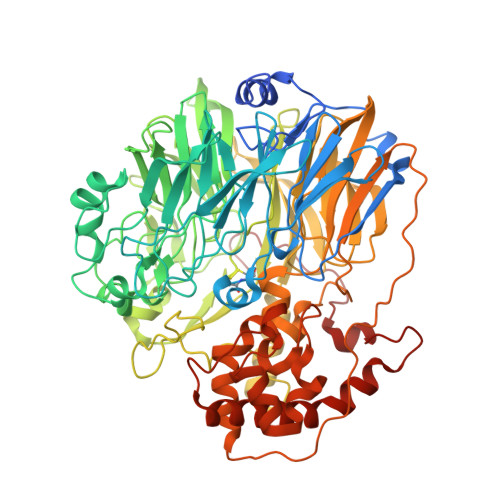
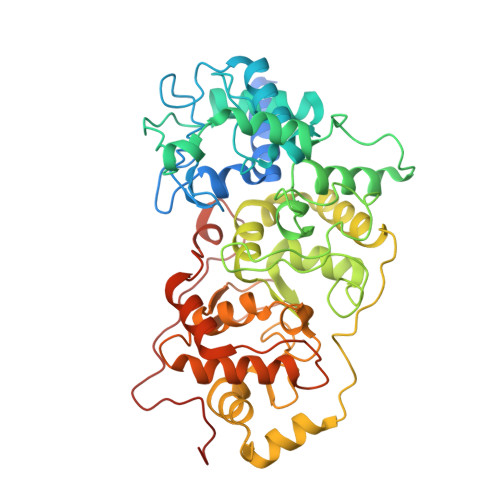
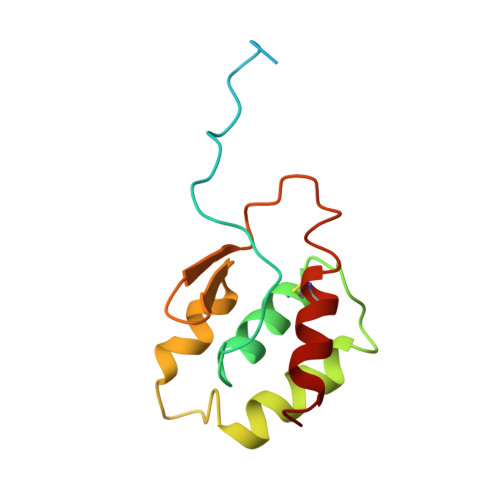
Experimental and Theoretical Insights into Bienzymatic Cascade for Mediatorless Bioelectrochemical Ethanol Oxidation with Alcohol and Aldehyde Dehydrogenases
Adachi, T., Miyata, T., Makino, F., Tanaka, H., Namba, K., Kano, K., Sowa, K., Kitazumi, Y., Shirai, O.(2023) ACS Catal 13: 7955-7965