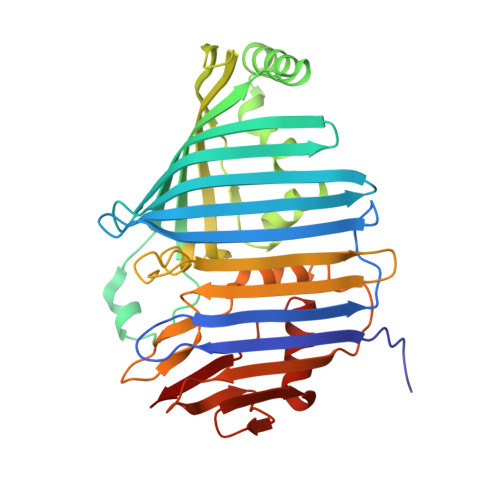
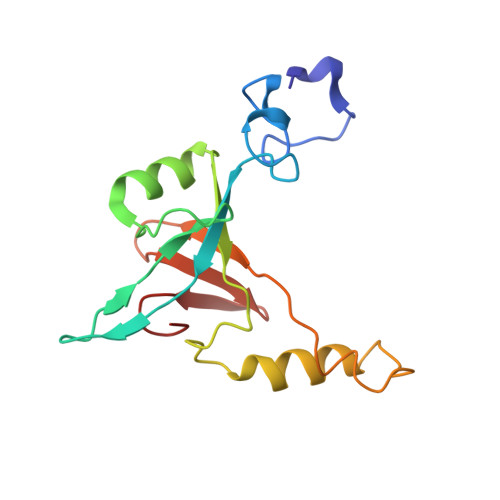
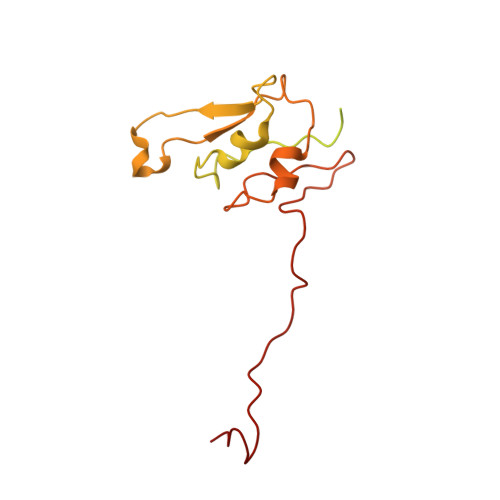
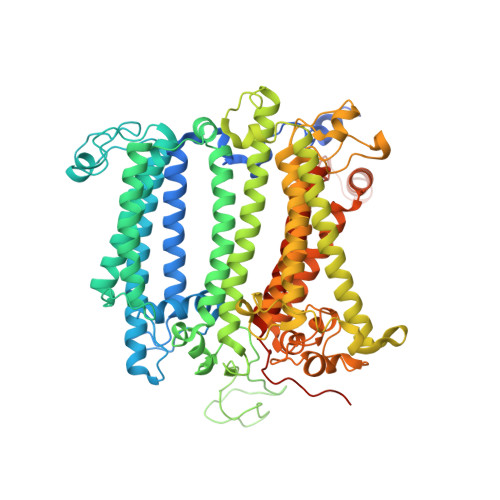
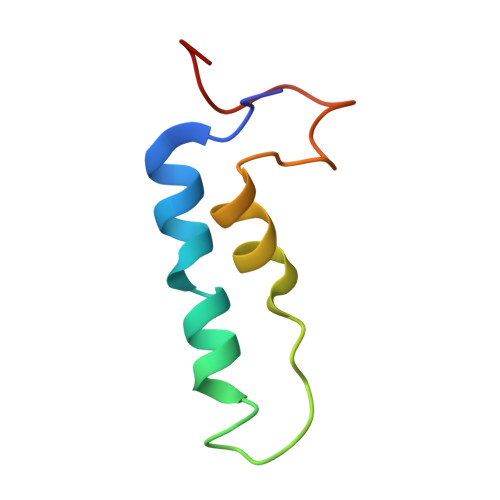
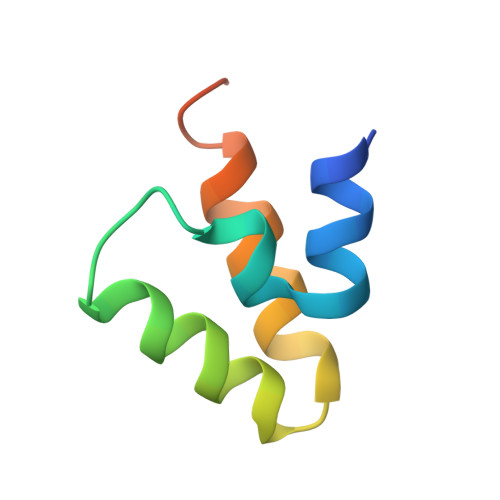
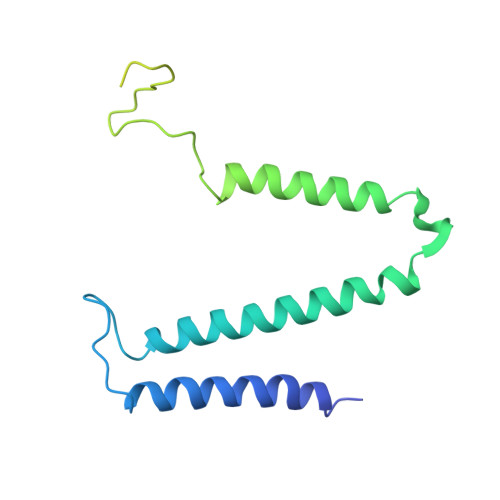
Cryo-electron microscopy structure of the intact photosynthetic light-harvesting antenna-reaction center complex from a green sulfur bacterium.
Chen, J.H., Wang, W., Wang, C., Kuang, T., Shen, J.R., Zhang, X.(2023) J Integr Plant Biol 65: 223-234
- PubMed: 36125941
- DOI: https://doi.org/10.1111/jipb.13367
- Primary Citation of Related Structures:
8GWA - PubMed Abstract:
The photosynthetic reaction center complex (RCC) of green sulfur bacteria (GSB) consists of the membrane-imbedded RC core and the peripheric energy transmitting proteins called Fenna-Matthews-Olson (FMO). Functionally, FMO transfers the absorbed energy from a huge peripheral light-harvesting antenna named chlorosome to the RC core where charge separation occurs. In vivo, one RC was found to bind two FMOs, however, the intact structure of RCC as well as the energy transfer mechanism within RCC remain to be clarified. Here we report a structure of intact RCC which contains a RC core and two FMO trimers from a thermophilic green sulfur bacterium Chlorobaculum tepidum at 2.9 Å resolution by cryo-electron microscopy. The second FMO trimer is attached at the cytoplasmic side asymmetrically relative to the first FMO trimer reported previously. We also observed two new subunits (PscE and PscF) and the N-terminal transmembrane domain of a cytochrome-containing subunit (PscC) in the structure. These two novel subunits possibly function to facilitate the binding of FMOs to RC core and to stabilize the whole complex. A new bacteriochlorophyll (numbered as 816) was identified at the interspace between PscF and PscA-1, causing an asymmetrical energy transfer from the two FMO trimers to RC core. Based on the structure, we propose an energy transfer network within this photosynthetic apparatus.
- College of Life Science, Zhejiang University, Hangzhou, 310058, China.
Organizational Affiliation: