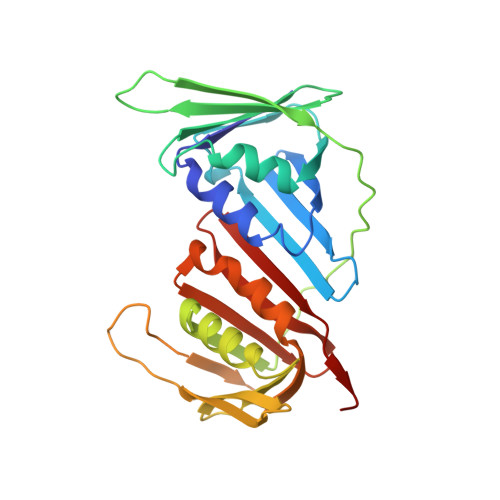
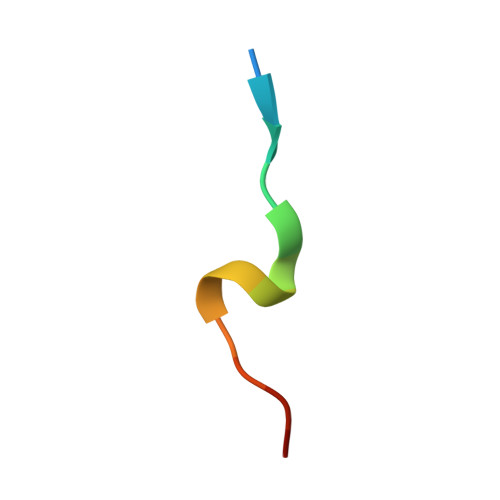
Towards a High-Affinity Peptidomimetic Targeting Proliferating Cell Nuclear Antigen from Aspergillus fumigatus.
Vandborg, B.C., Horsfall, A.J., Pederick, J.L., Abell, A.D., Bruning, J.B.(2023) J Fungi (Basel) 9
- PubMed: 37998903
- DOI: https://doi.org/10.3390/jof9111098
- Primary Citation of Related Structures:
8GJ5, 8GJF - PubMed Abstract:
Invasive fungal infections (IFIs) are prevalent in immunocompromised patients. Due to alarming levels of increasing resistance in clinical settings, new drugs targeting the major fungal pathogen Aspergillus fumigatus are required. Attractive drug targets are those involved in essential processes like DNA replication, such as proliferating cell nuclear antigens (PCNAs). PCNA has been previously studied in cancer research and presents a viable target for antifungals. Human PCNA interacts with the p21 protein, outcompeting binding proteins to halt DNA replication. The affinity of p21 for hPCNA has been shown to outcompete other associating proteins, presenting an attractive scaffold for peptidomimetic design. p21 has no A. fumigatus homolog to our knowledge, yet our group has previously demonstrated that human p21 can interact with A. fumigatus PCNA (afumPCNA). This suggests that a p21-based inhibitor could be designed to outcompete the native binding partners of afumPCNA to inhibit fungal growth. Here, we present an investigation of extensive structure-activity relationships between designed p21-based peptides and afumPCNA and the first crystal structure of a p21 peptide bound to afumPCNA, demonstrating that the A. fumigatus replication model uses a PIP-box sequence as the method for binding to afumPCNA. These results inform the new optimized secondary structure design of a potential peptidomimetic inhibitor of afumPCNA.
- Institute of Photonics and Advanced Sensing (IPAS), The University of Adelaide, Adelaide 5005, Australia.
Organizational Affiliation: