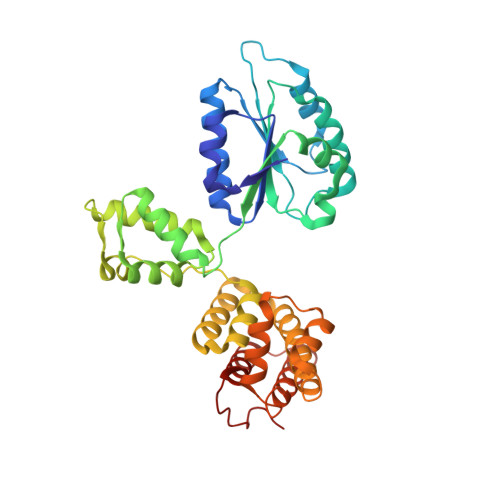
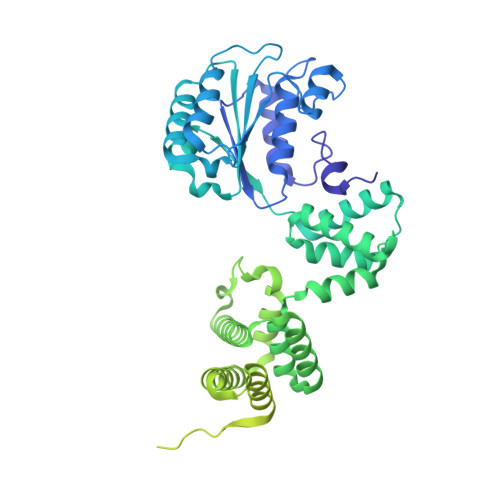
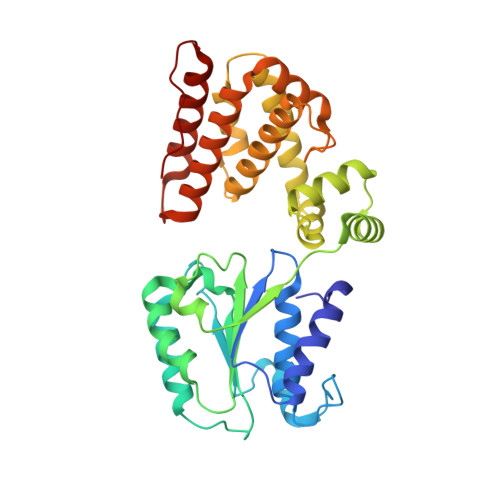
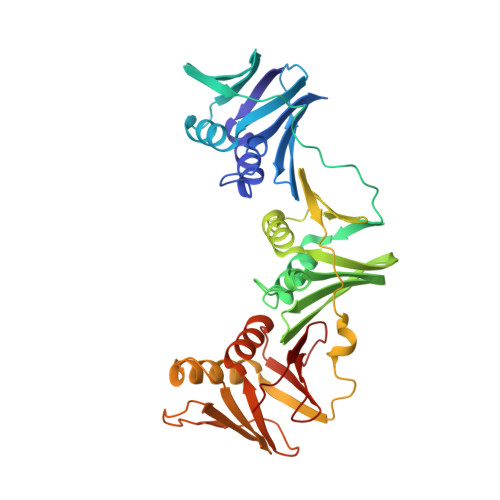
Structural characterisation of the complete cycle of sliding clamp loading in Escherichia coli.
Xu, Z.Q., Jergic, S., Lo, A.T.Y., Pradhan, A.C., Brown, S.H.J., Bouwer, J.C., Ghodke, H., Lewis, P.J., Tolun, G., Oakley, A.J., Dixon, N.E.(2024) Nat Commun 15: 8372-8372
- PubMed: 39333521
- DOI: https://doi.org/10.1038/s41467-024-52623-9
- Primary Citation of Related Structures:
8GIY, 8GIZ, 8GJ0, 8GJ1, 8GJ2, 8GJ3 - PubMed Abstract:
Ring-shaped DNA sliding clamps are essential for DNA replication and genome maintenance. Clamps need to be opened and chaperoned onto DNA by clamp loader complexes (CLCs). Detailed understanding of the mechanisms by which CLCs open and place clamps around DNA remains incomplete. Here, we present a series of six structures of the Escherichia coli CLC bound to an open or closed clamp prior to and after binding to a primer-template DNA, representing the most significant intermediates in the clamp loading process. We show that the ATP-bound CLC first binds to a clamp, then constricts to hold onto it. The CLC then expands to open the clamp with a gap large enough for double-stranded DNA to enter. Upon binding to DNA, the CLC constricts slightly, allowing clamp closing around DNA. These structures provide critical high-resolution snapshots of clamp loading by the E. coli CLC, revealing how the molecular machine works.
- Molecular Horizons and School of Chemistry and Molecular Bioscience, University of Wollongong, Wollongong, Australia. zhiqiang@uow.edu.au.
Organizational Affiliation: