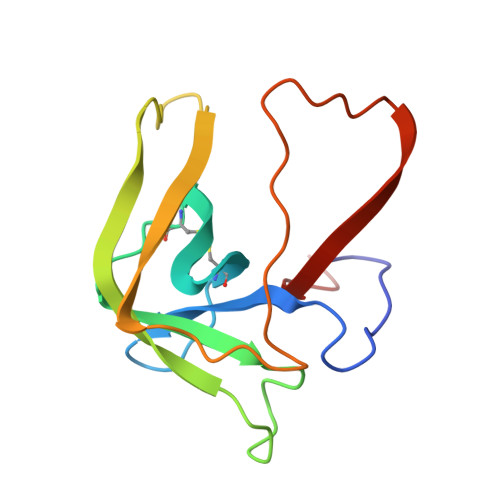
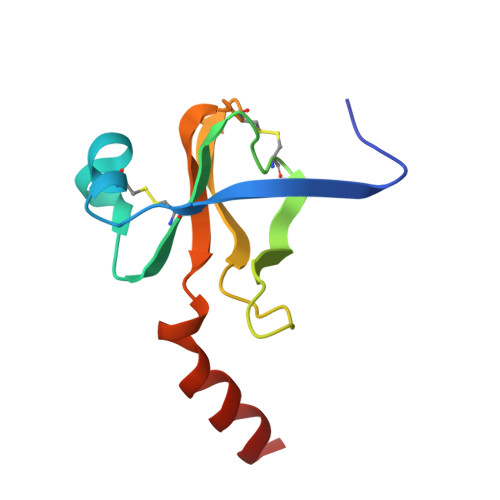
Gamma-chymotrypsin is a complex of alpha-chymotrypsin with its own autolysis products.
Harel, M., Su, C.T., Frolow, F., Silman, I., Sussman, J.L.(1991) Biochemistry 30: 5217-5225
- PubMed: 2036388
- DOI: https://doi.org/10.1021/bi00235a015
- Primary Citation of Related Structures:
8GCH - PubMed Abstract:
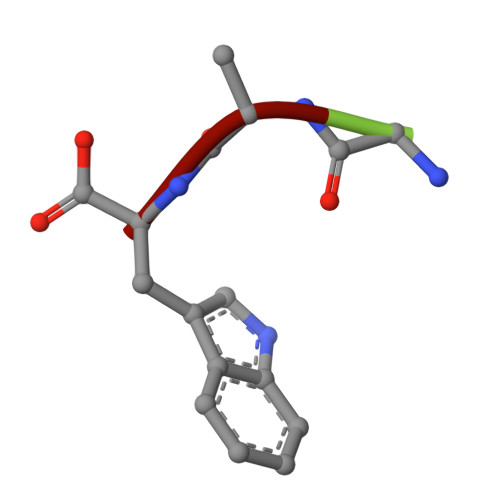
The determination of three separate gamma-chymotrypsin structures at different temperatures and resolutions confirmed the presence of electron density in the active site, which could be interpreted as an oligopeptide as had previously been suggested by Dixon and Matthews [(1989) Biochemistry 28, 7033-7038]. HPLC analyses of the enzyme before and after crystallization demonstrated the presence of a wide variety of oligopeptides in the redissolved crystal, most with COOH-terminal aromatic residues, as expected of the products of chymotrypsin cleavage, which appeared to arise from extensive autolysis of the enzyme under the crystallization conditions. The refined structures agree well with the conformation of both gamma-chymotrypsin and alpha-chymotrypsin. The electron density in the active site is thus interpreted as arising from a repertoire of autolysed oligopeptides produced concomitantly with crystallization. The COOH-terminal carbons of the polypeptide(s) display short contact distances (1.97, 2.47, and 2.13 A, respectively) to Ser195 O gamma in all three refined structures, but the electron density is not continuous between these two atoms in any of them. This suggests that some sequences are covalently bound as enzyme intermediates while others are noncovalently bound as enzyme-product complexes.
- Department of Structural Chemistry, Weizmann Institute of Science, Rehovot, Israel.
Organizational Affiliation: