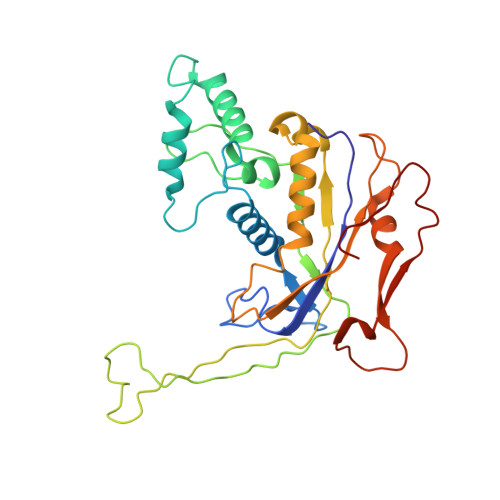
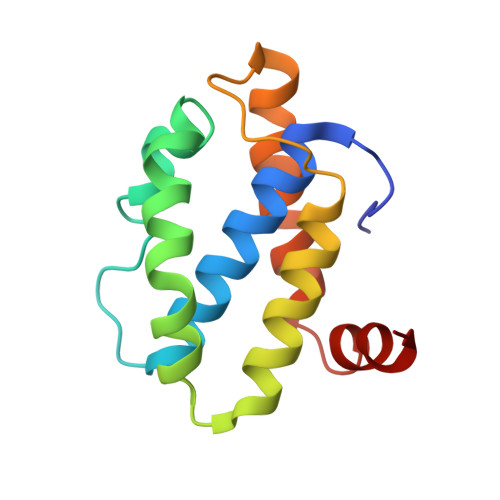
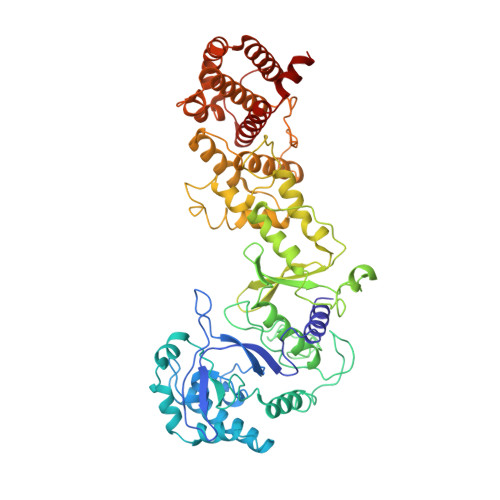
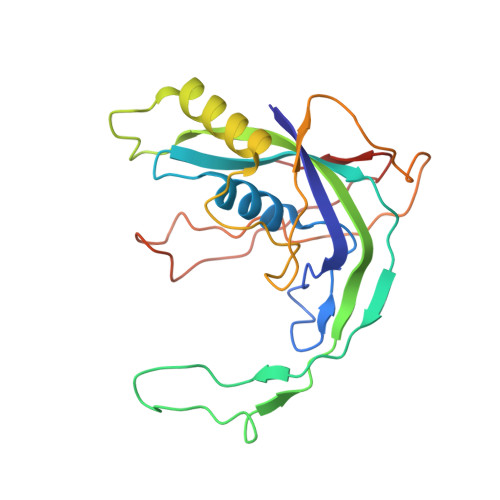
Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Hu, C., Myers, M.T., Zhou, X., Hou, Z., Lozen, M.L., Nam, K.H., Zhang, Y., Ke, A.(2024) Mol Cell 84: 463-475.e5
- PubMed: 38242128
- DOI: https://doi.org/10.1016/j.molcel.2023.12.034
- Primary Citation of Related Structures:
8G9S, 8G9T, 8G9U, 8GAF, 8GAM, 8GAN - PubMed Abstract:
Type I CRISPR-Cas systems utilize the RNA-guided Cascade complex to identify matching DNA targets and the nuclease-helicase Cas3 to degrade them. Among the seven subtypes, type I-C is compact in size and highly active in creating large-sized genome deletions in human cells. Here, we use four cryoelectron microscopy snapshots to define its RNA-guided DNA binding and cleavage mechanisms in high resolution. The non-target DNA strand (NTS) is accommodated by I-C Cascade in a continuous binding groove along the juxtaposed Cas11 subunits. Binding of Cas3 further traps a flexible bulge in NTS, enabling NTS nicking. We identified two anti-CRISPR proteins AcrIC8 and AcrIC9 that strongly inhibit Neisseria lactamica I-C function. Structural analysis showed that AcrIC8 inhibits PAM recognition through allosteric inhibition, whereas AcrIC9 achieves so through direct competition. Both Acrs potently inhibit I-C-mediated genome editing and transcriptional modulation in human cells, providing the first off-switches for type I CRISPR eukaryotic genome engineering.
- Department of Molecular Biology and Genetics, Cornell University, 253 Biotechnology Building, Ithaca, NY 14853, USA; Department of Biological Sciences, Faculty of Science; Department of Biochemistry, Precision Medicine Translational Research Programme (TRP), Yong Loo Lin School of Medicine, National University of Singapore, Singapore 117597, Singapore.
Organizational Affiliation: