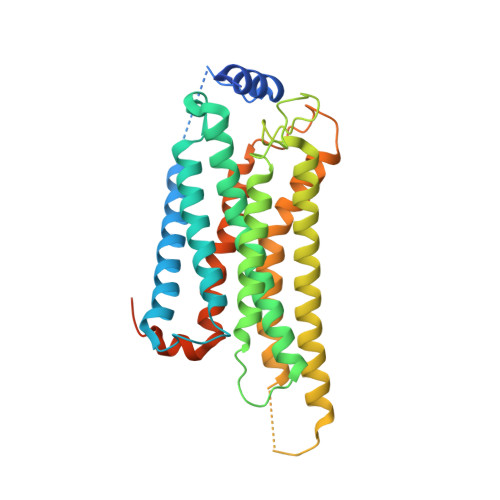
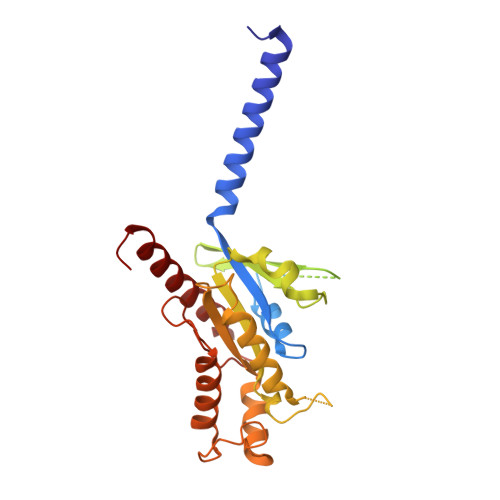
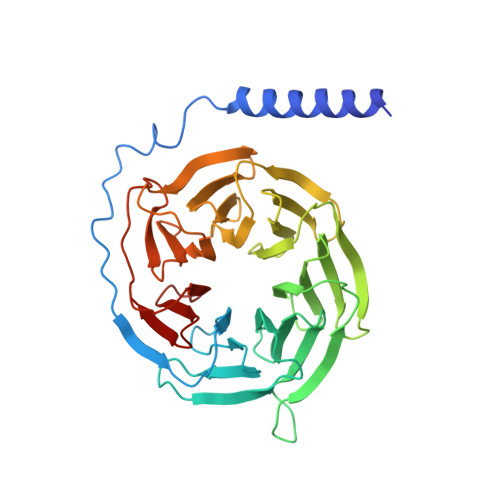
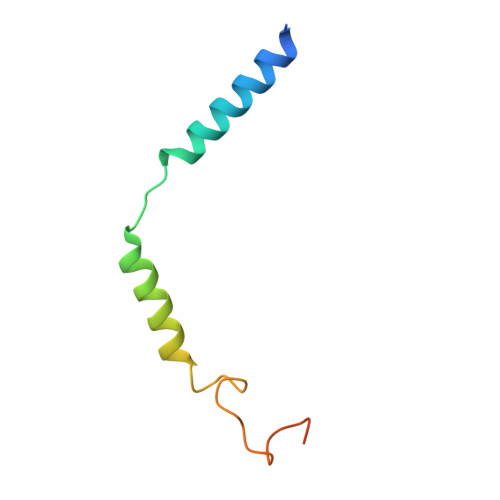
Transmembrane protein CD69 acts as an S1PR1 agonist.
Chen, H., Qin, Y., Chou, M., Cyster, J.G., Li, X.(2023) Elife 12
- PubMed: 37039481
- DOI: https://doi.org/10.7554/eLife.88204
- Primary Citation of Related Structures:
8G94 - PubMed Abstract:
The activation of Sphingosine-1-phosphate receptor 1 (S1PR1) by S1P promotes lymphocyte egress from lymphoid organs, a process critical for immune surveillance and T cell effector activity. Multiple drugs that inhibit S1PR1 function are in use clinically for the treatment of autoimmune diseases. Cluster of Differentiation 69 (CD69) is an endogenous negative regulator of lymphocyte egress that interacts with S1PR1 in cis to facilitate internalization and degradation of the receptor. The mechanism by which CD69 causes S1PR1 internalization has been unclear. Moreover, although there are numerous class A GPCR structures determined with different small molecule agonists bound, it remains unknown whether a transmembrane protein per se can act as a class A GPCR agonist. Here, we present the cryo-EM structure of CD69-bound S1PR1 coupled to the heterotrimeric G i complex. The transmembrane helix (TM) of one protomer of CD69 homodimer contacts the S1PR1-TM4. This interaction allosterically induces the movement of S1PR1-TMs 5-6, directly activating the receptor to engage the heterotrimeric G i . Mutations in key residues at the interface affect the interactions between CD69 and S1PR1, as well as reduce the receptor internalization. Thus, our structural findings along with functional analyses demonstrate that CD69 acts in cis as a protein agonist of S1PR1, thereby promoting G i -dependent S1PR1 internalization, loss of S1P gradient sensing, and inhibition of lymphocyte egress.
- Department of Molecular Genetics, The University of Texas Southwestern Medical Center, Dallas, United States.
Organizational Affiliation: