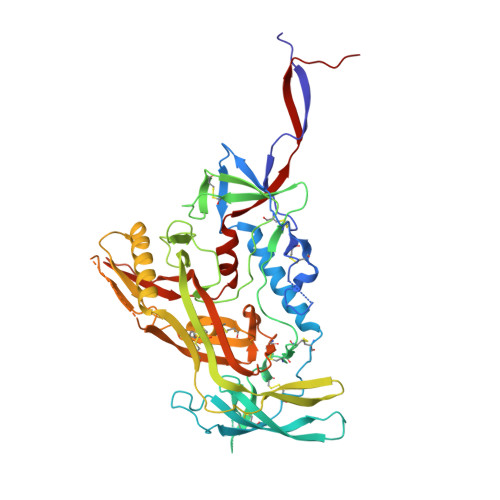
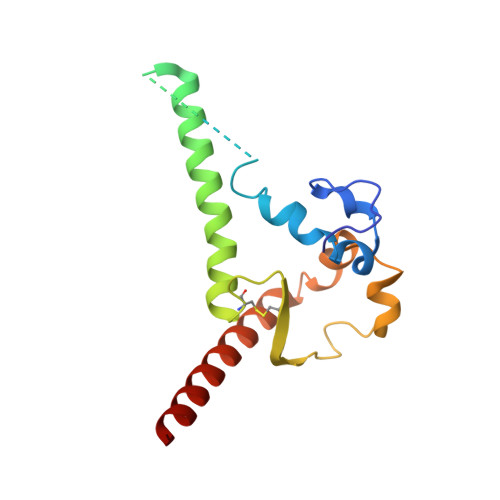
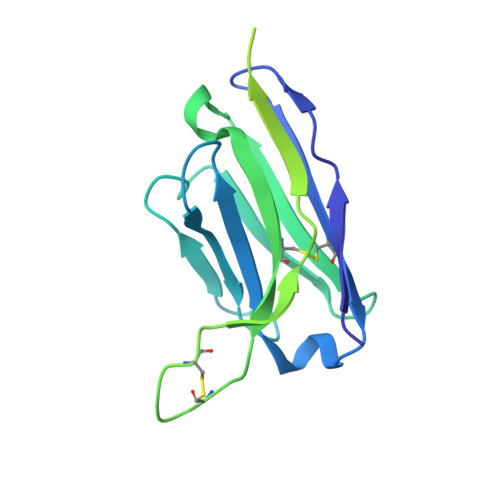
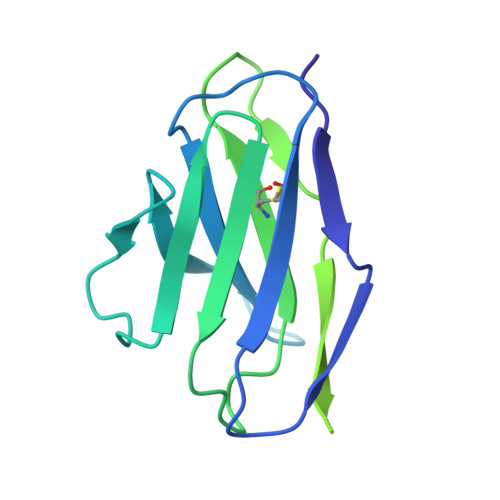
HIV-1 neutralizing antibodies elicited in humans by a prefusion-stabilized envelope trimer form a reproducible class targeting fusion peptide.
Wang, S., Matassoli, F., Zhang, B., Liu, T., Shen, C.H., Bylund, T., Johnston, T., Henry, A.R., Teng, I.T., Tripathi, P., Becker, J.E., Changela, A., Chaudhary, R., Cheng, C., Gaudinski, M., Gorman, J., Harris, D.R., Lee, M., Morano, N.C., Novik, L., O'Dell, S., Olia, A.S., Parchment, D.K., Rawi, R., Roberts-Torres, J., Stephens, T., Tsybovsky, Y., Wang, D., Van Wazer, D.J., Zhou, T., Doria-Rose, N.A., Koup, R.A., Shapiro, L., Douek, D.C., McDermott, A.B., Kwong, P.D.(2023) Cell Rep 42: 112755-112755
- PubMed: 37436899
- DOI: https://doi.org/10.1016/j.celrep.2023.112755
- Primary Citation of Related Structures:
8G4M, 8G4T - PubMed Abstract:
Elicitation of antibodies that neutralize the tier-2 neutralization-resistant isolates that typify HIV-1 transmission has been a long-sought goal. Success with prefusion-stabilized envelope trimers eliciting autologous neutralizing antibodies has been reported in multiple vaccine-test species, though not in humans. To investigate elicitation of HIV-1 neutralizing antibodies in humans, here, we analyze B cells from a phase I clinical trial of the "DS-SOSIP"-stabilized envelope trimer from strain BG505, identifying two antibodies, N751-2C06.01 and N751-2C09.01 (named for donor-lineage.clone), that neutralize the autologous tier-2 strain, BG505. Though derived from distinct lineages, these antibodies form a reproducible antibody class that targets the HIV-1 fusion peptide. Both antibodies are highly strain specific, which we attribute to their partial recognition of a BG505-specific glycan hole and to their binding requirements for a few BG505-specific residues. Prefusion-stabilized envelope trimers can thus elicit autologous tier-2 neutralizing antibodies in humans, with initially identified neutralizing antibodies recognizing the fusion-peptide site of vulnerability.
- Vaccine Research Center, National Institutes of Health, Bethesda, MD 20892, USA.
Organizational Affiliation: