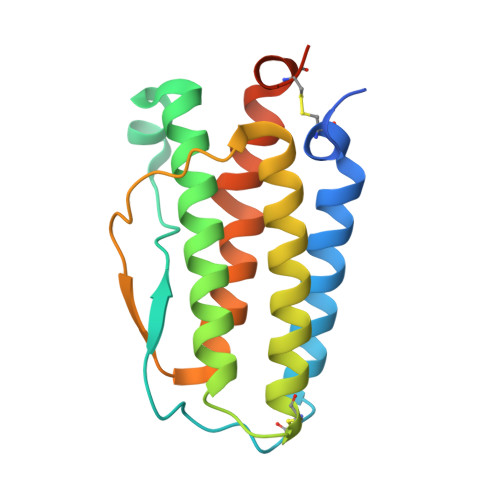
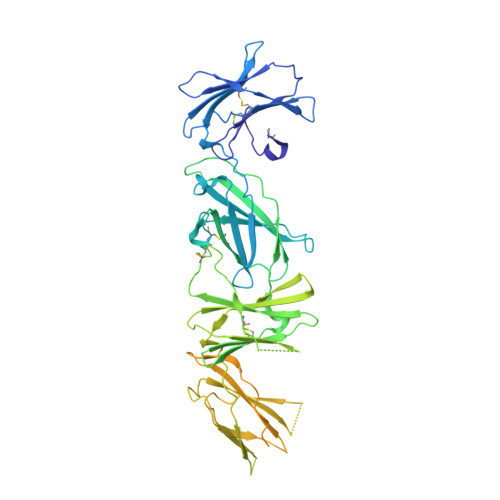
Structure of the thrombopoietin-MPL receptor complex is a blueprint for biasing hematopoiesis.
Tsutsumi, N., Masoumi, Z., James, S.C., Tucker, J.A., Winkelmann, H., Grey, W., Picton, L.K., Moss, L., Wilson, S.C., Caveney, N.A., Jude, K.M., Gati, C., Piehler, J., Hitchcock, I.S., Garcia, K.C.(2023) Cell 186: 4189-4203.e22
- PubMed: 37633268
- DOI: https://doi.org/10.1016/j.cell.2023.07.037
- Primary Citation of Related Structures:
8G04 - PubMed Abstract:
Thrombopoietin (THPO or TPO) is an essential cytokine for hematopoietic stem cell (HSC) maintenance and megakaryocyte differentiation. Here, we report the 3.4 Å resolution cryoelectron microscopy structure of the extracellular TPO-TPO receptor (TpoR or MPL) signaling complex, revealing the basis for homodimeric MPL activation and providing a structural rationalization for genetic loss-of-function thrombocytopenia mutations. The structure guided the engineering of TPO variants (TPO mod ) with a spectrum of signaling activities, from neutral antagonists to partial- and super-agonists. Partial agonist TPO mod decoupled JAK/STAT from ERK/AKT/CREB activation, driving a bias for megakaryopoiesis and platelet production without causing significant HSC expansion in mice and showing superior maintenance of human HSCs in vitro. These data demonstrate the functional uncoupling of the two primary roles of TPO, highlighting the potential utility of TPO mod in hematology research and clinical HSC transplantation.
- Department of Molecular and Cellular Physiology, Stanford University School of Medicine, Stanford, CA 94305, USA; Department of Structural Biology, Stanford University School of Medicine, Stanford, CA 94305, USA; Howard Hughes Medical Institute, Stanford University School of Medicine, Stanford, CA 94305, USA; Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama University, Okayama 700-8530, Japan. Electronic address: ntsutsumi@okayama-u.ac.jp.
Organizational Affiliation: