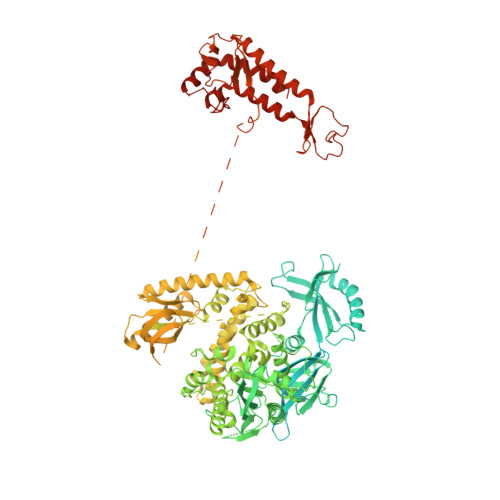
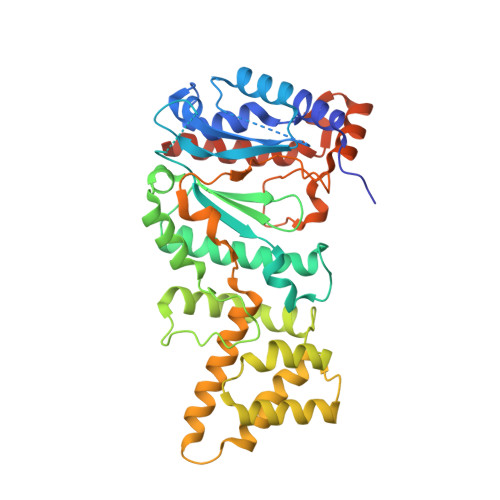
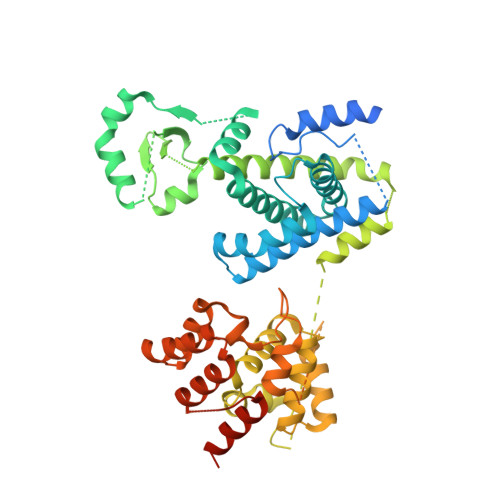
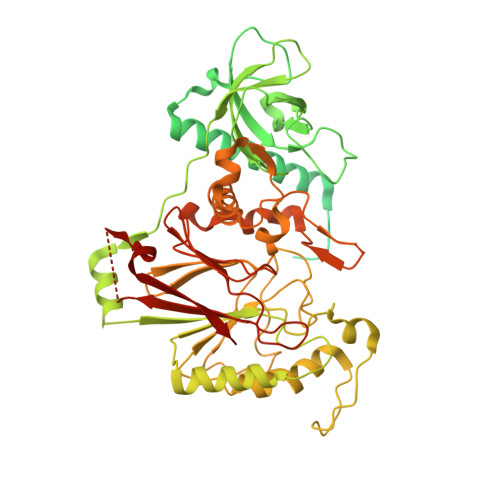
Molecular choreography of primer synthesis by the eukaryotic Pol alpha-primase.
Yuan, Z., Georgescu, R., Li, H., O'Donnell, M.E.(2023) Nat Commun 14: 3697-3697
- PubMed: 37344454
- DOI: https://doi.org/10.1038/s41467-023-39441-1
- Primary Citation of Related Structures:
8FOC, 8FOD, 8FOE, 8FOH, 8FOJ, 8FOK - PubMed Abstract:
The eukaryotic polymerase α (Pol α) synthesizes an RNA-DNA hybrid primer of 20-30 nucleotides. Pol α is composed of Pol1, Pol12, Primase 1 (Pri1), and Pri2. Pol1 and Pri1 contain the DNA polymerase and RNA primase activities, respectively. It has been unclear how Pol α hands over an RNA primer from Pri1 to Pol1 for DNA primer extension, and how the primer length is defined. Here we report the cryo-EM analysis of yeast Pol α in the apo, primer initiation, primer elongation, RNA primer hand-off from Pri1 to Pol1, and DNA extension states, revealing a series of very large movements. We reveal a critical point at which Pol1-core moves to take over the 3'-end of the RNA from Pri1. DNA extension is limited by a spiral motion of Pol1-core. Since both Pri1 and Pol1-core are flexibly attached to a stable platform, primer growth produces stress that limits the primer length.
- Department of Structural Biology, Van Andel Institute, Grand Rapids, MI, USA.
Organizational Affiliation: