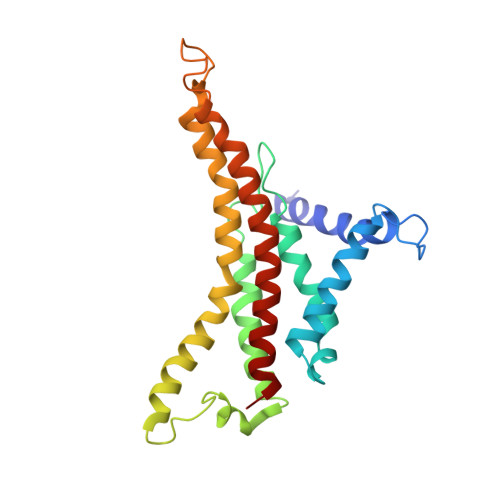
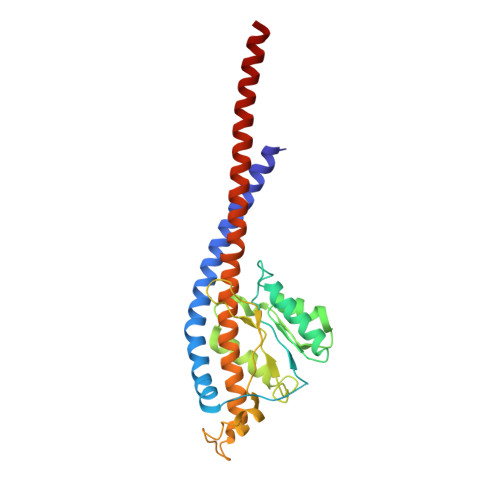
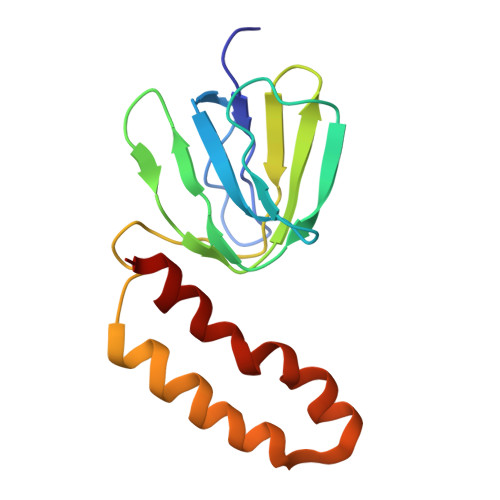
Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Sharma, S., Luo, M., Patel, H., Mueller, D.M., Liao, M.(2024) Nat Struct Mol Biol 31: 657-666
- PubMed: 38316880
- DOI: https://doi.org/10.1038/s41594-024-01219-4
- Primary Citation of Related Structures:
8F29, 8F39, 8FKJ, 8FL8 - PubMed Abstract:
Mitochondrial adenosine triphosphate (ATP) synthase uses the proton gradient across the inner mitochondrial membrane to synthesize ATP. Structural and single molecule studies conducted mostly at neutral or basic pH have provided details of the reaction mechanism of ATP synthesis. However, pH of the mitochondrial matrix is slightly acidic during hypoxia and pH-dependent conformational changes in the ATP synthase have been reported. Here we use single-particle cryo-EM to analyze the conformational ensemble of the yeast (Saccharomyces cerevisiae) ATP synthase at pH 6. Of the four conformations resolved in this study, three are reaction intermediates. In addition to canonical catalytic dwell and binding dwell structures, we identify two unique conformations with nearly identical positions of the central rotor but different catalytic site conformations. These structures provide new insights into the catalytic mechanism of the ATP synthase and highlight elastic coupling between the catalytic and proton translocating domains.
- Department of Cell Biology, Blavatnik Institute, Harvard Medical School, Boston, MA, USA. stuti.sharma@stonybrook.edu.
Organizational Affiliation: