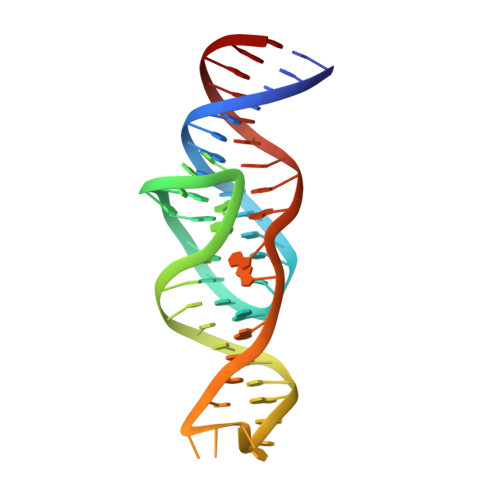
Intricate 3D architecture of a DNA mimic of GFP.
Passalacqua, L.F.M., Banco, M.T., Moon, J.D., Li, X., Jaffrey, S.R., Ferre-D'Amare, A.R.(2023) Nature 618: 1078-1084
- PubMed: 37344591
- DOI: https://doi.org/10.1038/s41586-023-06229-8
- Primary Citation of Related Structures:
8FHV, 8FHX, 8FHZ, 8FI0, 8FI1, 8FI2, 8FI7, 8FI8 - PubMed Abstract:
Numerous studies have shown how RNA molecules can adopt elaborate three-dimensional (3D) architectures 1-3 . By contrast, whether DNA can self-assemble into complex 3D folds capable of sophisticated biochemistry, independent of protein or RNA partners, has remained mysterious. Lettuce is an in vitro-evolved DNA molecule that binds and activates 4 conditional fluorophores derived from GFP. To extend previous structural studies 5,6 of fluorogenic RNAs, GFP and other fluorescent proteins 7 to DNA, we characterize Lettuce-fluorophore complexes by X-ray crystallography and cryogenic electron microscopy. The results reveal that the 53-nucleotide DNA adopts a four-way junction (4WJ) fold. Instead of the canonical L-shaped or H-shaped structures commonly seen 8 in 4WJ RNAs, the four stems of Lettuce form two coaxial stacks that pack co-linearly to form a central G-quadruplex in which the fluorophore binds. This fold is stabilized by stacking, extensive nucleobase hydrogen bonding-including through unusual diagonally stacked bases that bridge successive tiers of the main coaxial stacks of the DNA-and coordination of monovalent and divalent cations. Overall, the structure is more compact than many RNAs of comparable size. Lettuce demonstrates how DNA can form elaborate 3D structures without using RNA-like tertiary interactions and suggests that new principles of nucleic acid organization will be forthcoming from the analysis of complex DNAs.
- Laboratory of Nucleic Acids, National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD, USA.
Organizational Affiliation: