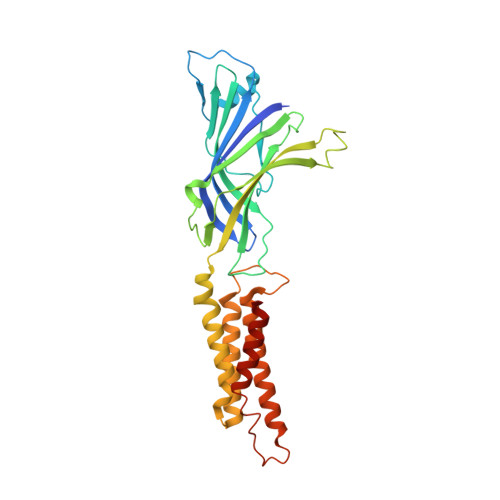
Lipid nanodisc scaffold and size alter the structure of a pentameric ligand-gated ion channel.
Dalal, V., Arcario, M.J., Petroff 2nd, J.T., Tan, B.K., Dietzen, N.M., Rau, M.J., Fitzpatrick, J.A.J., Brannigan, G., Cheng, W.W.L.(2024) Nat Commun 15: 25-25
- PubMed: 38167383
- DOI: https://doi.org/10.1038/s41467-023-44366-w
- Primary Citation of Related Structures:
8F32, 8F33, 8F34, 8F35, 8TWV, 8TWZ - PubMed Abstract:
Lipid nanodiscs have become a standard tool for studying membrane proteins, including using single particle cryo-electron microscopy (cryo-EM). We find that reconstituting the pentameric ligand-gated ion channel (pLGIC), Erwinia ligand-gated ion channel (ELIC), in different nanodiscs produces distinct structures by cryo-EM. The effect of the nanodisc on ELIC structure extends to the extracellular domain and agonist binding site. Additionally, molecular dynamic simulations indicate that nanodiscs of different size impact ELIC structure and that the nanodisc scaffold directly interacts with ELIC. These findings suggest that the nanodisc plays a crucial role in determining the structure of pLGICs, and that reconstitution of ion channels in larger nanodiscs may better approximate a lipid membrane environment.
- Department of Anesthesiology, Washington University School of Medicine, Saint Louis, MO, USA.
Organizational Affiliation: