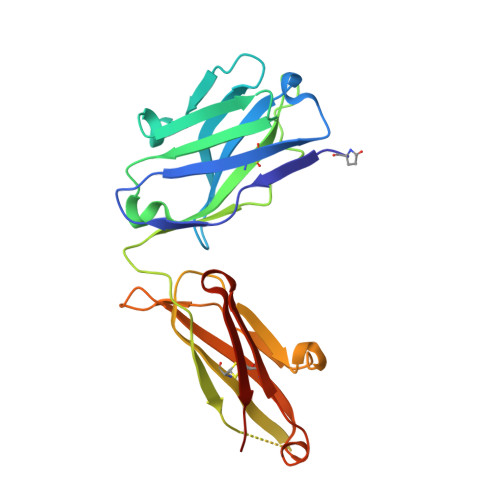
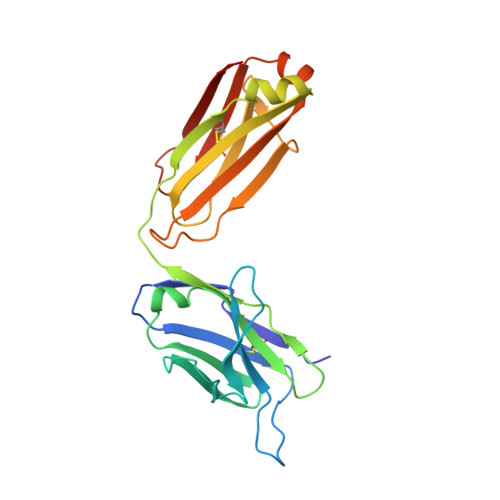
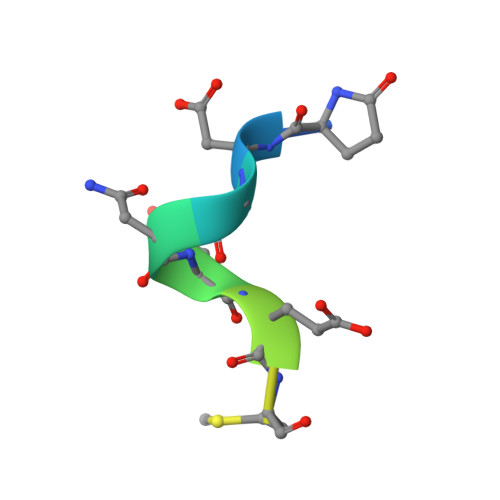
Structure-based engineering of a novel CD3 epsilon-targeting antibody for reduced polyreactivity.
Liu, C.Y., Ahonen, C.L., Brown, M.E., Zhou, L., Welin, M., Krauland, E.M., Pejchal, R., Widboom, P.F., Battles, M.B.(2023) MAbs 15: 2189974-2189974
- PubMed: 36991534
- DOI: https://doi.org/10.1080/19420862.2023.2189974
- Primary Citation of Related Structures:
8F0L - PubMed Abstract:
Bispecific antibodies continue to represent a growth area for antibody therapeutics, with roughly a third of molecules in clinical development being T-cell engagers that use an anti-CD3 binding arm. CD3 antibodies possessing cross-reactivity with cynomolgus monkey typically recognize a highly electronegative linear epitope at the extreme N-terminus of CD3 epsilon (CD3ε). Such antibodies have high isoelectric points and display problematic polyreactivity (correlated with poor pharmacokinetics for monospecific antibodies). Using insights from the crystal structure of anti-Hu/Cy CD3 antibody ADI-26906 in complex with CD3ε and antibody engineering using a yeast-based platform, we have derived high-affinity CD3 antibody variants with very low polyreactivity and significantly improved biophysical developability. Comparison of these variants with CD3 antibodies in the clinic (as part of bi- or multi-specifics) shows that affinity for CD3 is correlated with polyreactivity. Our engineered CD3 antibodies break this correlation, forming a broad affinity range with no to low polyreactivity. Such antibodies will enable bispecifics with improved pharmacokinetic and safety profiles and suggest engineering solutions that will benefit the large and growing sector of T-cell engagers.
- Adimab, LLC, NH, USA.
Organizational Affiliation: