Novel cofactor binding motifs, electron transfer and ion pumping mechanisms of the respiratory complex NQR
Juarez, O., Fuller, J.R.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Na(+)-translocating NADH-quinone reductase subunit A | 446 | Vibrio cholerae O395 | Mutation(s): 0 Gene Names: nqrA, VC_2295 EC: 7.2.1.1 | 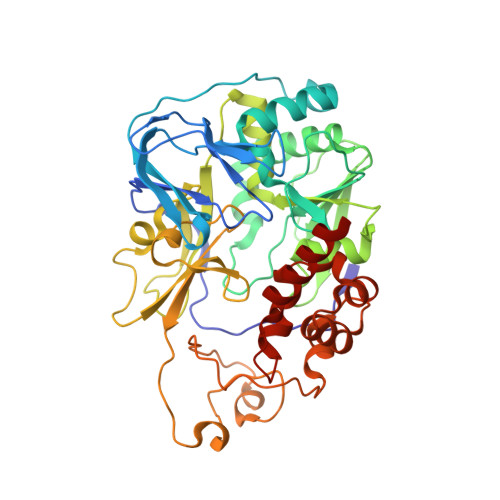 | |
UniProt | |||||
Find proteins for A5F5X1 (Vibrio cholerae serotype O1 (strain ATCC 39541 / Classical Ogawa 395 / O395)) Explore A5F5X1 Go to UniProtKB: A5F5X1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A5F5X1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Na(+)-translocating NADH-quinone reductase subunit B | 415 | Vibrio cholerae O395 | Mutation(s): 0 Gene Names: nqrB, VC_2294 EC: 7.2.1.1 Membrane Entity: Yes | 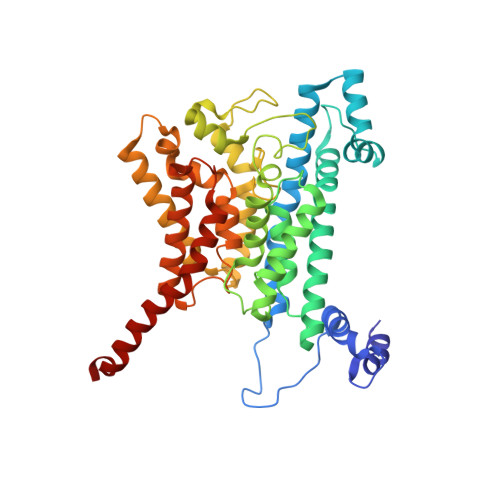 | |
UniProt | |||||
Find proteins for A5F5X0 (Vibrio cholerae serotype O1 (strain ATCC 39541 / Classical Ogawa 395 / O395)) Explore A5F5X0 Go to UniProtKB: A5F5X0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A5F5X0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Na(+)-translocating NADH-quinone reductase subunit C | 257 | Vibrio cholerae O395 | Mutation(s): 0 Gene Names: nqrC, VC_2293 EC: 7.2.1.1 Membrane Entity: Yes | 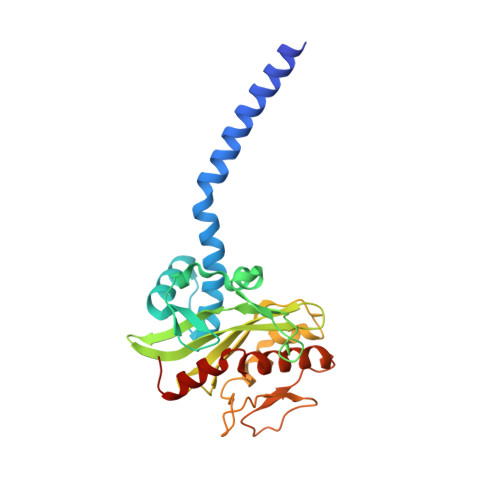 | |
UniProt | |||||
Find proteins for A5F5Y7 (Vibrio cholerae serotype O1 (strain ATCC 39541 / Classical Ogawa 395 / O395)) Explore A5F5Y7 Go to UniProtKB: A5F5Y7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A5F5Y7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Na(+)-translocating NADH-quinone reductase subunit D | 210 | Vibrio cholerae O395 | Mutation(s): 0 Gene Names: nqrD, VC_2292 EC: 7.2.1.1 Membrane Entity: Yes | 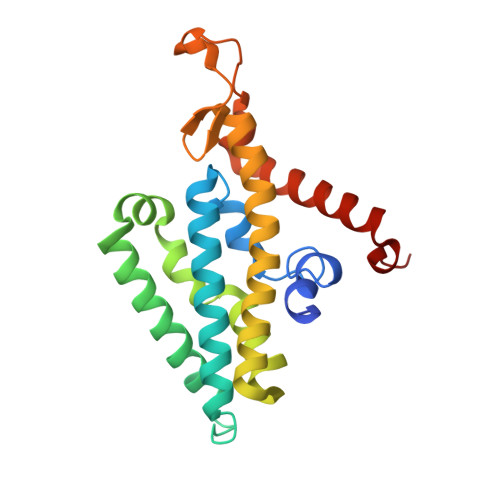 | |
UniProt | |||||
Find proteins for A5F5Y6 (Vibrio cholerae serotype O1 (strain ATCC 39541 / Classical Ogawa 395 / O395)) Explore A5F5Y6 Go to UniProtKB: A5F5Y6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A5F5Y6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Na(+)-translocating NADH-quinone reductase subunit E | 198 | Vibrio cholerae O395 | Mutation(s): 0 Gene Names: nqrE, VC_2291 EC: 7.2.1.1 Membrane Entity: Yes |  | |
UniProt | |||||
Find proteins for A5F5Y5 (Vibrio cholerae serotype O1 (strain ATCC 39541 / Classical Ogawa 395 / O395)) Explore A5F5Y5 Go to UniProtKB: A5F5Y5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A5F5Y5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Na(+)-translocating NADH-quinone reductase subunit F | 408 | synthetic construct | Mutation(s): 0 Gene Names: nqrF, VC_2290 EC: 7.2.1.1 Membrane Entity: Yes |  | |
UniProt | |||||
Find proteins for A5F5Y4 (Vibrio cholerae serotype O1 (strain ATCC 39541 / Classical Ogawa 395 / O395)) Explore A5F5Y4 Go to UniProtKB: A5F5Y4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A5F5Y4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| FMN (Subject of Investigation/LOI) Query on FMN | G [auth B], J [auth C] | FLAVIN MONONUCLEOTIDE C17 H21 N4 O9 P FVTCRASFADXXNN-SCRDCRAPSA-N |  | ||
| RBF (Subject of Investigation/LOI) Query on RBF | H [auth B] | RIBOFLAVIN C17 H20 N4 O6 AUNGANRZJHBGPY-SCRDCRAPSA-N |  | ||
| UQ1 (Subject of Investigation/LOI) Query on UQ1 | I [auth B] | UBIQUINONE-1 C14 H18 O4 SOECUQMRSRVZQQ-UHFFFAOYSA-N |  | ||
| FES (Subject of Investigation/LOI) Query on FES | K [auth E] | FE2/S2 (INORGANIC) CLUSTER Fe2 S2 NIXDOXVAJZFRNF-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | RELION | 4.0beta2 |
| MODEL REFINEMENT | PHENIX | |
| MODEL REFINEMENT | ISOLDE | |
| MODEL REFINEMENT | Rosetta |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) | United States | AI151152 |