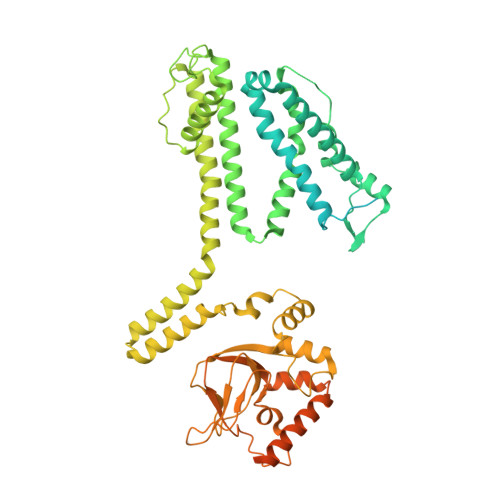
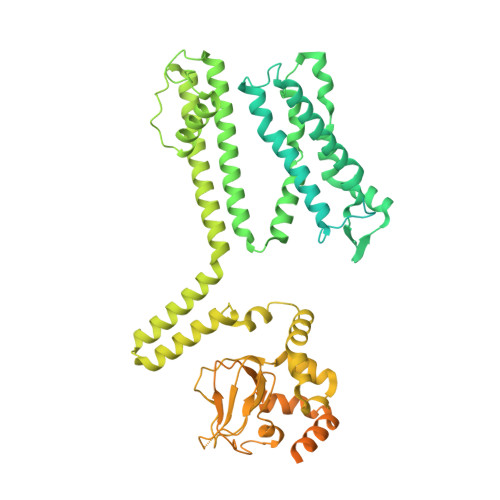
Conformational trajectory of allosteric gating of the human cone photoreceptor cyclic nucleotide-gated channel.
Hu, Z., Zheng, X., Yang, J.(2023) Nat Commun 14: 4284-4284
- PubMed: 37463923
- DOI: https://doi.org/10.1038/s41467-023-39971-8
- Primary Citation of Related Structures:
8ETP, 8EU3, 8EUC, 8EV8, 8EV9, 8EVA, 8EVB, 8EVC - PubMed Abstract:
Cyclic nucleotide-gated (CNG) channels transduce chemical signals into electrical signals in sensory receptors and neurons. They are activated by cGMP or cAMP, which bind to the cyclic nucleotide-binding domain (CNBD) to open a gate located 50-60 Å away in the central cavity. Structures of closed and open vertebrate CNG channels have been solved, but the conformational landscape of this allosteric gating remains to be elucidated and enriched. Here, we report structures of the cGMP-activated human cone photoreceptor CNGA3/CNGB3 channel in closed, intermediate, pre-open and open states in detergent or lipid nanodisc, all with fully bound cGMP. The pre-open and open states are obtained only in the lipid nanodisc, suggesting a critical role of lipids in tuning the energetic landscape of CNGA3/CNGB3 activation. The different states exhibit subunit-unique, incremental and distinct conformational rearrangements that originate in the CNBD, propagate through the gating ring to the transmembrane domain, and gradually open the S6 cavity gate. Our work illustrates a spatial conformational-change wave of allosteric gating of a vertebrate CNG channel by its natural ligand and provides an expanded framework for studying CNG properties and channelopathy.
- Department of Biological Sciences, Columbia University, New York, NY, 10027, USA.
Organizational Affiliation: