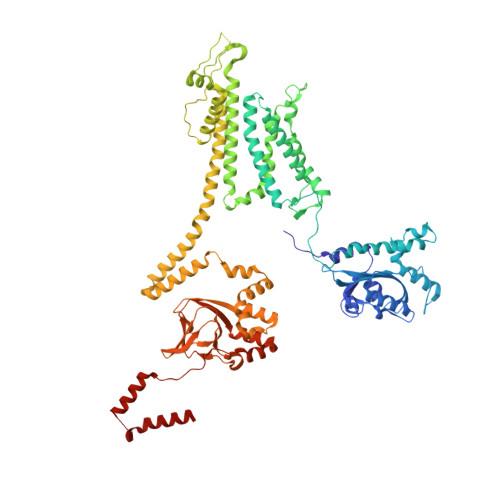
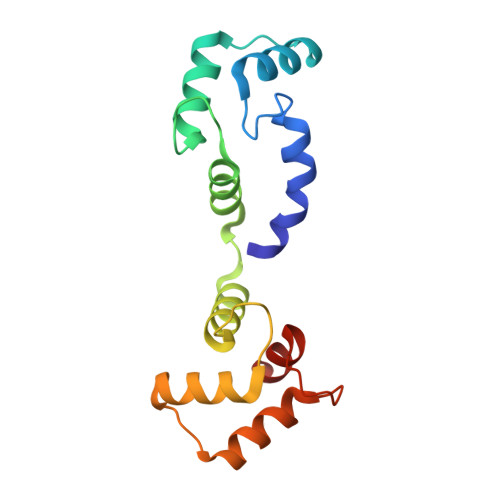
Voltage-sensor movements in the Eag Kv channel under an applied electric field.
Mandala, V.S., MacKinnon, R.(2022) Proc Natl Acad Sci U S A 119: e2214151119-e2214151119
- PubMed: 36331999
- DOI: https://doi.org/10.1073/pnas.2214151119
- Primary Citation of Related Structures:
8EOW, 8EP0, 8EP1 - PubMed Abstract:
Voltage-dependent ion channels regulate the opening of their pores by sensing the membrane voltage. This process underlies the propagation of action potentials and other forms of electrical activity in cells. The voltage dependence of these channels is governed by the transmembrane displacement of the positive charged S4 helix within their voltage-sensor domains. We use cryo-electron microscopy to visualize this movement in the mammalian Eag voltage-dependent potassium channel in lipid membrane vesicles with a voltage difference across the membrane. Multiple structural configurations show that the applied electric field displaces S4 toward the cytoplasm by two helical turns, resulting in an extended interfacial helix near the inner membrane leaflet. The position of S4 in this down conformation is sterically incompatible with an open pore, thus explaining how movement of the voltage sensor at hyperpolarizing membrane voltages locks the pore shut in this kind of voltage-dependent K + (K v ) channel. The structures solved in lipid bilayer vesicles detail the intricate interplay between K v channels and membranes, from showing how arginines are stabilized deep within the membrane and near phospholipid headgroups, to demonstrating how the channel reshapes the inner leaflet of the membrane itself.
- Laboratory of Molecular Neurobiology and Biophysics, The Rockefeller University, New York, NY, 10065.
Organizational Affiliation: