Structures of Ric-8B in complex with G alpha protein folding clients reveal isoform specificity mechanisms.
Papasergi-Scott, M.M., Kwarcinski, F.E., Yu, M., Panova, O., Ovrutsky, A.M., Skiniotis, G., Tall, G.G.(2023) Structure 31: 553-564.e7
- PubMed: 36931277
- DOI: https://doi.org/10.1016/j.str.2023.02.011
- Primary Citation of Related Structures:
8EL7, 8EL8 - PubMed Abstract:
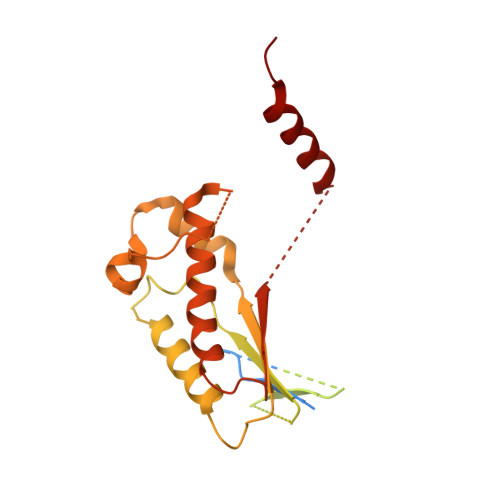
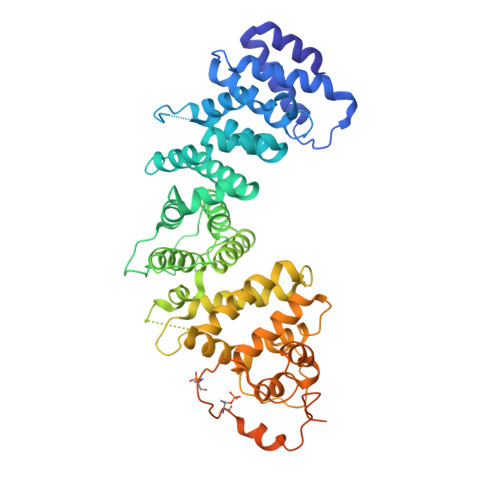
Mammalian Ric-8 proteins act as chaperones to regulate the cellular abundance of heterotrimeric G protein α subunits. The Ric-8A isoform chaperones Gαi/o, Gα12/13, and Gαq/11 subunits, while Ric-8B acts on Gαs/olf subunits. Here, we determined cryoelectron microscopy (cryo-EM) structures of Ric-8B in complex with Gαs and Gαolf, revealing isoform differences in the relative positioning and contacts between the C-terminal α5 helix of Gα within the concave pocket formed by Ric-8 α-helical repeat elements. Despite the overall architectural similarity with our earlier structures of Ric-8A complexed to Gαq and Gαi1, Ric-8B distinctly accommodates an extended loop found only in Gαs/olf proteins. The structures, along with results from Ric-8 protein thermal stability assays and cell-based Gαolf folding assays, support a requirement for the Gα C-terminal region for binding specificity, and highlight that multiple structural elements impart specificity for Ric-8/G protein binding.
- Department of Molecular and Cellular Physiology, Stanford University School of Medicine, Stanford, CA 94305, USA.
Organizational Affiliation: