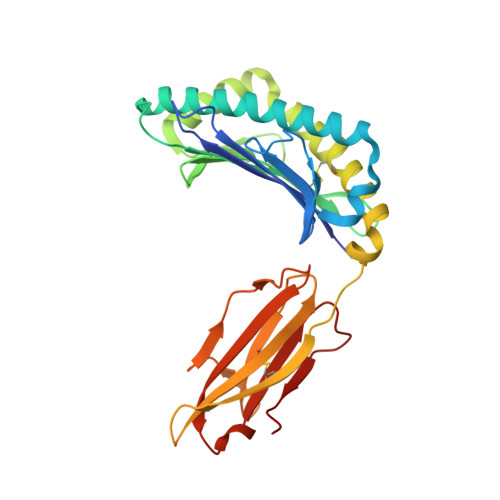
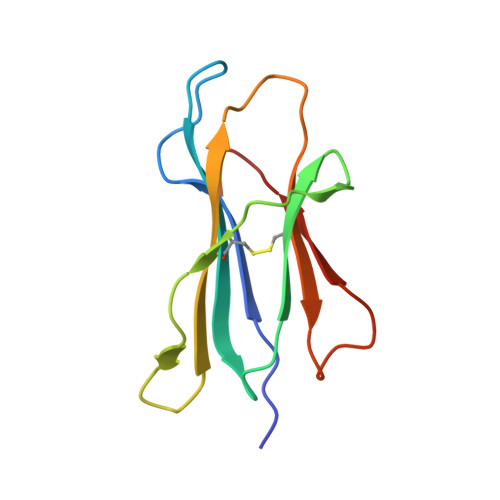
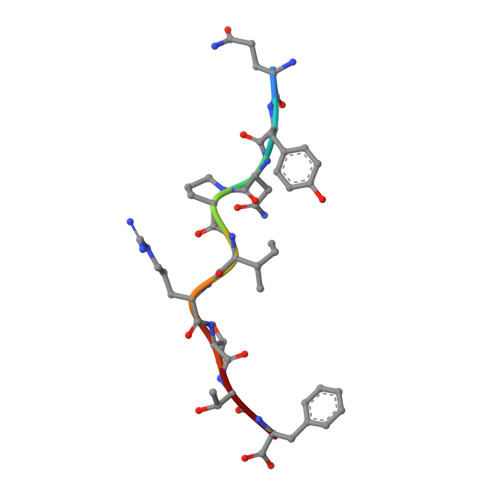
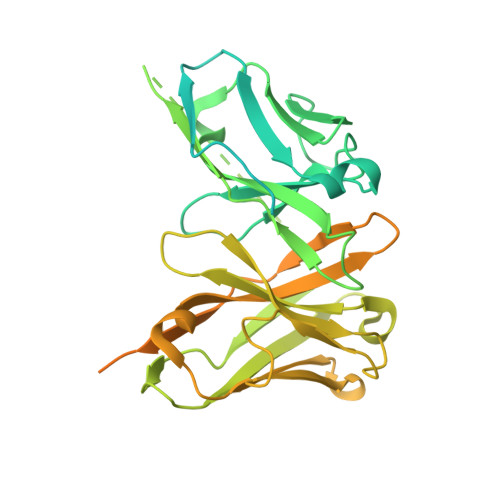
Structural principles of peptide-centric chimeric antigen receptor recognition guide therapeutic expansion.
Sun, Y., Florio, T.J., Gupta, S., Young, M.C., Marshall, Q.F., Garfinkle, S.E., Papadaki, G.F., Truong, H.V., Mycek, E., Li, P., Farrel, A., Church, N.L., Jabar, S., Beasley, M.D., Kiefel, B.R., Yarmarkovich, M., Mallik, L., Maris, J.M., Sgourakis, N.G.(2023) Sci Immunol 8: eadj5792-eadj5792
- PubMed: 38039376
- DOI: https://doi.org/10.1126/sciimmunol.adj5792
- Primary Citation of Related Structures:
8EK5, 8SBK, 8SBL - PubMed Abstract:
Peptide-centric chimeric antigen receptors (PC-CARs) recognize oncoprotein epitopes displayed by cell-surface human leukocyte antigens (HLAs) and offer a promising strategy for targeted cancer therapy. We have previously developed a PC-CAR targeting a neuroblastoma-associated PHOX2B peptide, leading to robust tumor cell lysis restricted by two common HLA allotypes. Here, we determine the 2.1-angstrom crystal structure of the PC-CAR-PHOX2B-HLA-A*24:02-β 2 m complex, which reveals the basis for antigen-specific recognition through interactions with CAR complementarity-determining regions (CDRs). This PC-CAR adopts a diagonal docking mode, where interactions with both conserved and polymorphic HLA framework residues permit recognition of multiple HLA allotypes from the A9 serological cross-reactive group, covering a combined global population frequency of up to 46.7%. Biochemical binding assays, molecular dynamics simulations, and structural and functional analyses demonstrate that high-affinity PC-CAR recognition of cross-reactive pHLAs necessitates the presentation of a specific peptide backbone, where subtle structural adaptations of the peptide are critical for high-affinity complex formation, and CAR T cell killing. Our results provide a molecular blueprint for engineering CARs with optimal recognition of tumor-associated antigens in the context of different HLAs, while minimizing cross-reactivity with self-epitopes.
- Center for Computational and Genomic Medicine and Department of Pathology and Laboratory Medicine, Children's Hospital of Philadelphia, Philadelphia, PA, USA.
Organizational Affiliation: