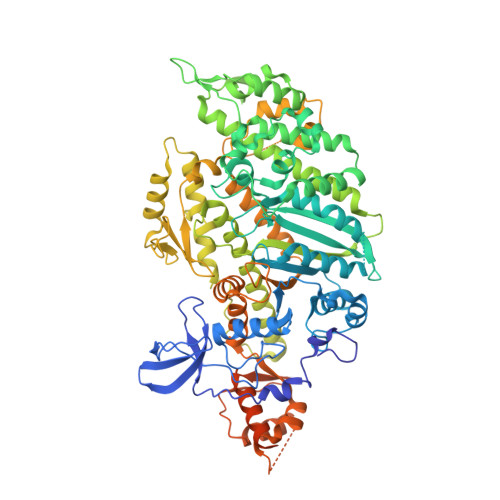
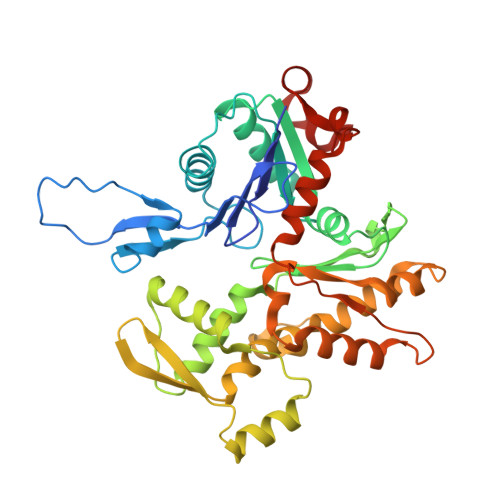
Conformational changes linked to ADP release from human cardiac myosin bound to actin-tropomyosin.
Doran, M.H., Rynkiewicz, M.J., Rassici, D., Bodt, S.M.L., Barry, M.E., Bullitt, E., Yengo, C.M., Moore, J.R., Lehman, W.(2023) J Gen Physiol 155
- PubMed: 36633586
- DOI: https://doi.org/10.1085/jgp.202213267
- Primary Citation of Related Structures:
8EFD, 8EFE, 8EFH - PubMed Abstract:
Following binding to the thin filament, β-cardiac myosin couples ATP-hydrolysis to conformational rearrangements in the myosin motor that drive myofilament sliding and cardiac ventricular contraction. However, key features of the cardiac-specific actin-myosin interaction remain uncertain, including the structural effect of ADP release from myosin, which is rate-limiting during force generation. In fact, ADP release slows under experimental load or in the intact heart due to the afterload, thereby adjusting cardiac muscle power output to meet physiological demands. To further elucidate the structural basis of this fundamental process, we used a combination of cryo-EM reconstruction methodologies to determine structures of the human cardiac actin-myosin-tropomyosin filament complex at better than 3.4 Å-resolution in the presence and in the absence of Mg2+·ADP. Focused refinements of the myosin motor head and its essential light chains in these reconstructions reveal that small changes in the nucleotide-binding site are coupled to significant rigid body movements of the myosin converter domain and a 16-degree lever arm swing. Our structures provide a mechanistic framework to understand the effect of ADP binding and release on human cardiac β-myosin, and offer insights into the force-sensing mechanism displayed by the cardiac myosin motor.
- School of Medicine, Department of Physiology and Biophysics, Boston University , Boston, MA, USA.
Organizational Affiliation: